Abstract
Small extracellular vesicles (sEVs), also referred as exosomes, have emerged as valuable indicators of cancer progression and response to treatment. They offer prospective targets for therapeutic interventions as well as insightful information about the fundamental mechanisms underlying the development of cancer. sEVs have garnered significant attention as a useful tool for liquid biopsies used in non-invasive cancer diagnosis. We discussed their potential in predicting treatment outcomes, monitoring disease progression, and classifying cancer stages and subtypes. sEVs can also shed light on how resistance to several cancer treatments, such as drug resistance, radiation resistance, chemotherapy resistance, and immunotherapy resistance develops. sEV-based cancer diagnostics have initiated clinical trials, underscoring their potential clinical value. Additionally, significant progress has been made in the development of techniques for isolating and enriching sEVs, enabling the sensitive and efficient detection of sEV proteins and nucleic acids. These advancements have resulted in enhanced sensitivity and specificity, facilitating the identification of biomarkers with low expression levels. In conclusion, sEV biomarkers offer significant potential for the diagnosis and monitoring of cancer. The utilization of sEVs in liquid biopsies presents a non-invasive method for acquiring tumour-specific information. Ongoing research and advancements in sEV-based diagnostics and therapeutics are crucial for unlocking the complete potential of sEV biomarkers in clinical settings.
Introduction
Small extracellular vesicles (sEVs) released by diverse cell types have emerged as promising biomarkers for diagnosis and monitoring of cancer. These EVs promote intercellular communication by transferring bioactive molecules, such as proteins, nucleic acids, and lipids.[1] A comprehensive understanding of the biogenesis of EVs is essential to comprehend their functional significance and unlock their potential as diagnostic tools in cancer.
EVs are classified into different subtypes based on their biogenesis pathways, with sEVs being the most extensively studied subtype.[2] sEVs originate from the endosomal pathway and are formed through a complex series of intracellular processes. Biogenesis of sEVs begins with the inward budding of the plasma membrane, leading to the formation of early endosomes. These early endosomes mature into multivesicular bodies (MVBs) through the inward budding of their limiting membrane.[3] Subsequently, MVBs can either fuse with lysosomes for degradation or undergo exocytosis, releasing their intraluminal vesicles as sEVs into the extracellular space.[4]
The cargo carried by sEVs is highly diverse and reflects the molecular composition of the parent cells.[5] This cargo encompasses various proteins, such as membrane transporters, signalling molecules, and receptors, as along with different types of nucleic acids, such as messenger RNA (mRNA), microRNA (miRNA), and long non-coding RNA (lncRNA).[6, 7, 8] The selective packaging of these molecules into sEVs suggests a tightly regulated sorting process during their biogenesis.[9,10]
The ability of sEVs to transfer their cargo to recipient cells has led to growing interest in their role in cancer progression and therapeutic resistance. sEVs derived from cancer cells can modulate the tumour microenvironment,[11] promote angiogenesis,[12] facilitate metastasis,[13] and contribute to immune evasion.[14] Moreover, the specific cargo carried by cancer cell-derived sEVs can reflect the molecular characteristics of the tumour,[15] providing valuable information for cancer diagnosis and monitoring.
Before the 1970s, surgery stood as the primary rational cancer treatment, making a significant milestone in oncology. This era laid the groundwork for the subsequent development of radiation therapy and anticancer chemotherapy. The period spanning from the 1970s to 2023 witnessed a remarkable expansion in cancer therapeutics, with the introduction of therapies such as immune checkpoint inhibitors, pharmacological hormone therapies, and chimeric antigen receptor T cell therapy. These advanced treatments enhanced the efficacy of cancer management. These therapies are positioned to retain their essential roles in cancer treatment moving forward. Furthermore, advancements in early detection methodologies hold promise in revolutionizing patient care by enabling interventions at earlier disease stages, potentially intercepting metastatic progression.[16]
In this review, we delve into the potential of sEV biomarkers in various aspects and also discuss the utility of sEVs in liquid biopsy for predicting treatment outcomes, monitoring disease progression, and identifying resistance mechanisms. Additionally, we highlight recent advancements in sEVs isolation and enrichment techniques, as well as novel methods for detecting sEV proteins and nucleic acids. By integrating sEV biomarkers into clinical practice, we envision a transformative impact on cancer diagnostics and an overall enhancement of patient care.
Liquid biopsies offer a convenient alternative to tissue biopsies,[17] enabling multiple sampling possible throughout a patient’s treatment. These liquid biopsies encompass various biomarkers, including circulating tumour cells,[18] cell-free DNA[19] and sEVs.[20] Over the past decade, circulating tumour DNA (ctDNA) was one of the most important components of liquid biopsy techniques. However, a significant challenge lies in the small fraction of ctDNA amidst the abundant background of normal cfDNA.[21] Among these biomarkers, sEVs stand out as particularly useful due to their higher abundance in circulation and increased stability, making them a good candidate for liquid biopsy applications. The distinct biological origins of cfDNA and EVs introduce difference in the representation of tumour heterogeneity, allowing differentiation between EV-DNA and cfDNA.[22] The membranous structures of sEVs provide a protective shield,[23] preventing the enclosed molecules from physical degradation. sEVs derived from cancer cells have demonstrated significant potential as biomarkers for detecting and monitoring diseases since they can be found in a variety of bodily fluids, such as blood, mucus, urine, and bronchial fluid, and sEVs derived from different cancer types carry a unique set of biomarkers (Figure 1). The non-invasive nature of acquiring cancer-derived sEVs enables sequential sampling of patients, offering valuable insights for early diagnosis, identifying cancer recurrence, assessing drug response, and stratifying patients across a wide range of cancer types. Advances in nanotechnology-driven biosensors has ushered in a new era of diagnostics, characterized by enhanced capabilities including high throughput analysis, minimal sample requirements, and cost-efficient detection of sEV biomarkers. The ongoing refinement and innovation of biosensor technologies may facilitate integration of sEV detection into clinical environments.[24] SEV detection has the potential to greatly assist in clinical decision-making processes and improve patient outcomes.

sEV biomarkers associated with several prevalent types of cancer. sEVs derived from cancer cells have emerged as a promising tool in the field of cancer liquid biopsy. They carry unique sets of proteins and nucleic acids that exhibit distinct expression patterns compared to non-cancerous cells. The identification and analysis of these exosomal biomarkers offer opportunities to develop precise diagnostic and prognostic tests. sEV: small extracellular vesicles.
sEVs as biomarkers for cancer liquid biopsy
Several studies have investigated the potential of utilizing specific sEV protein biomarkers for diagnosing cancer using patients’ samples. Promising sEV biomarkers have been identified for pancreatic cancer. A cell surface proteoglycan, glypican-1 (GPC1) is enriched in sEVs. GPC1 can be detected in serum of patients with pancreatic cancer, demonstrating an area under the curve (AUC) of 1.0 and a sensitivity and specificity of 100%.[25] However, GPC1 is not exclusive to pancreatic cancer cells. It has been reported that colorectal cancer cells also overexpress GPC1.[26] This indicates that GPC1 alone may not be sufficient as a standalone biomarker for pancreatic cancer, and the combination of several protein biomarkers may be necessary to achieve both sensitive and specific detection of pancreatic cancer. A detection panel consisting of sEV GPC1, sEV cluster of differentiation 82 (CD82) and serum carbohydrate antigen 19-9 (CA19-9) exhibit a promising diagnostic result of AUC of 0.942 to effectively differentiate healthy individuals from pancreatic cancer patients.[27] It worth considering the inclusion of multiple biomarkers that are found to be overexpressed in pancreatic cancer cells in a diagnostic test. For example, miRNA-10b,[28] mir-155,[29] mir-125b-5p[30] have been identified as overexpressed in pancreatic cancer and may serve as valuable additions for a more comprehensive assessment of the disease.
Epithelial cellular adhesion molecule (EpCAM) is of interest in cancer research due to its altered expression in various types of epithelial cells. The overexpression of EpCAM is also detected in other cancer types, such as breast cancer,[31] ovarian cancer[32] and prostate cancer.[33] This highlights the necessity to include multiple biomarkers for increasing the accuracy of diagnostic tests. Biomarkers such as sEV FRα, sEV CD24, and sEV EpCAM have been employed to differentiate ovarian cancer patients from the control group. While FRα levels are nearly undetectable in control samples and substantially lower in sEVs from ovarian cancer, CD24 and EpCAM are both well-studied biomarkers. For sEV CD24, sEV EpCAM, and sEV FRα, the AUCs are 1.00, 1.00, and 0.995 respectively.[34] The levels of three biomarkers can be taken into consideration together when diagnosing ovarian cancer. For accurate metastatic breast cancer diagnosis, eight sEV biomarkers are used, which includes CA 15-3, CA 125, carcinoembryonic antigen (CEA), human epidermal growth factor receptor 2 (HER2), epidermal growth factor receptor (EGFR), prostate-specific membrane antigen (PSMA), EpCAM and vascular endothelial growth factor (VEGF). The combination of these biomarkers demonstrates a high accuracy with an AUPRC of 0.9912 in distinguishing breast cancer patients from healthy control utilising sEVs.[35] There are still limited studies on diagnosing cancer from different cancer types due to the scarcity of exclusive biomarkers specifically expressed in a cancer type. Combining multiple biomarkers to diagnose cancer provides a more accurate result.
Tumour-derived sEVs actively communicate with the surrounding microenvironment through the expression of specific biomarkers. It is of utmost importance to identify these biomarkers, as they can serve a variety of purposes including predicting overall survival, assessing the risk of recurrence, classifying cancer stages and subtypes, and evaluating treatment resistance.
Predict outcomes and monitor progression
sEV biomarkers provide valuable insights into disease progression and survival rates in HCC. HCC patients with larger tumours or at later TNM stage have been found to exhibit lower blood sEV miRNA-638 levels, and those with these conditions additionally displayed poorer three- and five-year survival rates.[36] This suggests the role of sEV miR-638 as a circulating cancer biomarker to predict poor prognosis for HCC patients. It has also been discovered that sEV PD-L1 contributes to survival prediction. Shorter progression-free survival and overall survival are independently predicted by a rise in sEV PD-L1.[37] The progression of cancer can be monitored by tracking changes in sEV biomarker profiles overtime. SEV biomarkers can also be used to determine the likelihood of cancer recurring after initial treatment. For example, significant differences in miR-718 expression were observed in serum sEVs of HCC cases with recurrence following liver transplantation compared to those without recurrence.[38] This facilitates the identification of patients who require liver transplantation and aids in predicting HCC recurrence after surgery. sEV lncRNAs display disease-specific expression patterns, making them useful biomarkers for evaluating recurrences. To differentiate patients with recurrent colorectal cancer from those without recurrence, a 5-sEV lncRNAs panel was created.[39] Another study also utilized serum sEV lncRNAs in breast cancer for recurrence prediction.[40] The execution of an aggressive follow-up approach for patients who are at high risk of recurrence can be justified by stratifying patients based on their risk of recurrence.
Classify stages and subtypes
sEVs contains biomarkers which contribute to the staging of cancer by providing information about the tumor’s size, invasiveness, and spread. This information aids in determining the appropriate treatment approach and prognosis for patients. Plasma sEV Sox2ot has been shown to correlated with TNM stage. Sox2ot is crucial for inducing EMT and stem cell-like characteristics, and sEVs with a high concentration of Sox2ot can facilitate tumour invasion and metastasis.[41] Another biomarker, sEV TGF-B1, has shown a strong correlation with TNM. Patients with advanced gastric cancer, defined as TNM stages 2, 3, or 4, have demonstrated increased levels of sEV TGF-B1 compared to patients with stage 1 disease.[42] This indicates that monitoring the expression level of sEV biomarkers can be a useful tool in classifying the TNM stage.
sEV biomarkers also play a crucial role in classifying cancer into different subtypes. In the context of breast cancer subtypes, patients with HER2− and HR+ tumours have better prognoses than those with more aggressive triple-negative (ER−PR−HER2−; TNBC) or HR−HER2+ malignancies.[43] sEV miRNAs have been reported in these subtypes, with miR-335, miR-422a, and miR-628 showing significant differences between TNBC and HER2-positive individuals. The AUC values for miR-335, miR-422a, and miR-628 are of 0.737, 0.655, and 0.759 respectively. These sEV miRNAs have a sensitivity of 65% and 68% and a specificity of 84% and 81%, respectively, for differentiating between TNBC and HER2-positive individuals.[44] The classification utilizing sEVs can assist in tailoring treatment strategies to specific cancer subtypes.
Assess resistance in cancer
Chemotherapy, radiation, immunotherapy, and surgical excision are commonly used treatment modalities. Nonetheless, it is not uncommon for cancer patients to develop primary or acquired medication resistance, and emerging evidence suggests that sEVs may play a role in the dissemination of drug resistance. It was discovered that cisplatin-resistant cells and stomach cancer cells have elevated levels of sEV circ-0063526. This Cir-0063526 has been shown to be packaged in sEVs and delivered to sensitive cells, thereby promoting resistance to cisplatin. In patients with gastric cancer, high expression of sEV circ-0063526 has been associated with a poor response to cisplatin treatment.[45] Similarly, differential expression of several microRNAs (miR-425-3p, miR-1273h, miR-4755-5p, miR-9-5p, miR-146a-5p, and miR-215-5p) has been observed, with the highest fold change in platinum-resistant non-small cell lung cancer (NSCLC) patients when compared to platinum-sensitive NSCLC patients. In patients with NSCLC, high miR-425-3p has been identified as a powerful prognostic biomarker for low responsiveness to platinum-based chemotherapy.[46]
More than half of cancer patients use radiotherapy to treat localized cancer, relieve symptoms, or slow disease progression. Nonetheless, radioresistance continues to be the primary cause of radiation failure. A comparison between radioresistant and radiosensitive NSCLC patients revealed that sEV miR-96 was significantly overexpressed in the radioresistant group. An AUC value of 0.7496 was obtained for sEV miR-96’s radioresistance diagnostic capability, suggesting its potential as a useful biomarker for distinguishing patients with radioresistant NSCLC from those with radiosensitive NSCLC.[47]
Recently, immunotherapy has gained popularity as a treatment for cancer, encompassing various strategies such as immune system modulators, cancer vaccines, and immune checkpoint inhibitors. sEVs have been discovered to be a useful biomarker for choosing the most suitable patients for immunotherapy. sEV PD-L1 expression has been shown to be higher in patients receiving immune checkpoint inhibitor treatment. Its expression can be used to predict an overall survival of NSCLC patients. Patients who demonstrated a fold change in sEV PD-L1 expression equivalent to or higher than 1.86 exhibited a higher progression-free survival rate.[48] This suggests that sEV PD-L1 expression can help identifying NSCLC patients who are likely to benefit from immune checkpoint inhibitor therapy and have a more favourable prognosis. Moreover, a prognostic and diagnostic model using two sEV-derived genes, MYL6B and THOC2, has been developed. High expression of these genes was associated with greater expression patterns of immune checkpoint genes, such as PD-1, B7H, CTLA4, and TIM3, in patients with HCC.[49] Patients whose immune checkpoint expression is higher may likely to benefit from immunotherapy as it can boost the immune response resulting in greater therapeutic benefits.
Translation of sEVs in clinical trials
In recent years, there has been significant progress in the translation of sEVs for cancer diagnosis, leading to the initiation of clinical trials. These trials are designed to evaluate the potential utility of sEV biomarkers in detecting and monitoring various types of cancer. Clinical trials investigating sEV biomarkers aim to develop sensitive and specific diagnostic tools that can aid in early cancer detection, monitor treatment response, and track disease progression. The unique properties of sEVs, such as their stability in various body fluids (e.g., blood, urine), make them attractive candidates for liquid biopsies. Table 1 showcases several ongoing clinical trials that are actively studying sEVs as diagnostic and prognostic markers. These trials are specifically aimed at confirming the reliability of sEV nucleic acids and proteins as reliable markers. However, it is important to note that clinical reports and data analysis are pending publication. These clinical trials are expected to pave the way for the development of noninvasive, easily accessible, and cost-effective diagnostic tools for various types of cancer.
Clinical trials utilising sEV biomarkers.
| Title | ID | Dates | Purpose | Conditions diseases or |
|---|---|---|---|---|
| Development of a prognostic and predictive biomarker for locally advanced breast cancer patients treated with neoadjuvant chemotherapy using sEVs | NCT05955521 | Study start date: May 1, 2021 Estimated study completion date: July 1, 2028 | Prognosis | Triple negative breast cancer and HER2-postive breast cancer |
| Clinical relevance of detecting molecular abnormalities in glial tumor sEVs | NCT06116903 | Study start date: December 15, 2023 Estimated study completion date: December 15, 2025 | Diagnosis | Glioma |
| Interrogation of sEV-mediated intercellular signaling in patients with pancreatic cancer | NCT02393703 | Study start date: March 2015 Estimated study completion date: March 2025 | Prognosis | Pancreatic cancer and benign pancreatic disease |
| Molecular profiling of sEVs in tumor- draining vein of early-staged lung cancer (ExOnSite-Pro) | NCT04939324 | Study Start Date: June 21, 2021 Estimated study completion date: June 6, 2024 | Diagnosis | Non-small-cell lung cancer |
| A companion diagnostic study to develop circulating sEVs as predictive biomarkers for the response to immunotherapy in renal cell carcinoma | NCT05705583 | Study start date: January 1, 2023 Estimated study completion date: December 31, 2025 | Prognosis | Renal cell carcinoma |
| A prospective, multicenter cohort Study of urinary sEV lncRNAs for preoperative diagnosis of lymphatic metastasis in patients with bladder cancer | NCT05270174 | Study start date: June 1, 2023 Estimated study completion date: August 1, 2025 | Diagnosis | Bladder cancer |
| Study of sEVs in monitoring patients with sarcoma (EXOSARC) | NCT03800121 | Study start date: November 19, 2018 Estimated study completion date: November 19, 2025 | Prognosis | Sarcoma |
| A retrospective study to compare biomarker expression of sEVs derived from peripheral blood and primary lung cancer drainage pulmonary blood in lung cancer patients | NCT05587114 | Study Start Date: October 13, 2022 Estimated study completion date: December 31, 2025 | Diagnosis | Lung cancer |
| Early detection of pancreatic cancer: prospective study | NCT06388967 | Study start date: March 15, 2023 Estimated study completion date: November 21, 2025 | Diagnosis | Pancreatic cancer |
| A sEV-based liquid biopsy for the differential diagnosis of primary liver cancer | NCT06342414 | Study start date: March 15, 2024 Estimated study completion date: March 15, 2025 | Diagnosis | Hepatocellular carcinoma |
| Early detection of stomach cancer with a liquid biopsy based on exosomal micro-RNA | NCT06342427 | Study start date: March 15, 2023 Estimated study completion date: March 15, 2025 | Diagnosis | Gastric cancer |
Note: The table provides information on the clinical title, ID, cancer type, purpose, and dates of the clinical trials. These trials focus on utilising sEV biomarkers to develop diagnostic or prognostic test for cancer. sEV: small extracellular vesicles.
While genetic profiling in human tumour analysis is increasingly used for enhancing cancer diagnosis, these methodologies come with limitations. For example, RNA sequencing fails to identify variants located in noncoding DNA regions,[50] the production of short reads by next-generation sequencing techniques can pose challenges when sequencing genomes with complex repetitive regions. [51] Researchers envision a future where personalized cancer management is facilitated through the analysis of information carried by sEVs. This could result in earlier detection of cancer, enabling timely intervention, more effective treatment strategies tailored to individual patients, and ultimately improved patient outcomes.
New methods for sEV isolation and enrichment
While sEV biomarkers hold promise for clinical use, there are several limitations that need to be addressed before their widespread implementation. One of the primary challenges is the high throughput isolation of sEVs. Currently, there is a lack of standardized and efficient methods for isolating sEVs from various body fluids in large quantities. Existing isolation techniques often suffer from low yield and variability, which can affect the reliability and reproducibility of results.
Precipitation, size-based separation, ultracentrifugation, and immunoaffinity are some of the conventional techniques widely used to isolate sEVs. Ultracentrifugation is a commonly employed technique, which applies strong centrifugal forces to the sample, separating sEVs from other particles according to their size and density. The procedure involves pre-processing the sample to pellet larger particles, debris, and cells at lower speed, followed by higher speed ultracentrifugation to pellet sEVs.[52] Size exclusion chromatography separates particles according to their hydrodynamic size as they pass through a porous stationary phase. It offers mild conditions for separation, reducing potential harm to sEVs and preserving their integrity.[53] Precipitation polyethylene glycol (PEG) is another often employed method. By adding a high PEG concentration to the sample, sEVs aggregate and precipitate, allowing for further processing and purification.[54] However, these conventional isolation methods are not only tedious and costly, but also limited to low-throughput applications and require specialised instruments. In recent years, emerging methods and technologies have addressed these limitations, enabling the rapid and convenient enrichment of sEVs (Figure 2).

New methods for sEV isolation and enrichment. The figure shows four new exosome isolation and enrichment methods. Size-based isolation separates sEVs based on their size and large particle are excluded. Magnetic based isolation captures sEVs using magnetic beads conjugated with antibodies specific to exosome surface markers. Acoustic based isolation isolates sEVs using acoustic waves without the need of labelling. Electro-deposition separates sEVs via applying electric field across a sample and sEVs migrate to the oppositely charged electrode due to their negative charge. sEV: small extracellular vesicles.
Size-based isolation
Deterministic lateral displacement (DLD) is a technique employed in microfluidics for the separation of particles within the nanometre to micrometre size range. It utilizes a series of bifurcations in the laminar flow pattern created by an array of regularly spaced pillars. This innovative method enables the efficient separation of particles from biological samples.[55] Larger vesicles were laterally displaced across the array and collected at a side channel, while smaller vesicles flew out of the array in a zigzag pattern, thereby achieving the collection of sEVs. This resulted in the production of a nanoscale DL that can separate particles between 20 and 110 nm. This method demonstrates the size sorting of sEVs and enables fast colloid sorting in a continuous flow with single-particle resolution, paving the way for on-chip separation and diagnostics.[56] The use of nanoscale DLD also has been proved to successfully isolate extracellular vesicles from serum and urine samples.[57] Using double linked harmonic oscillations, EXODUS, an ultrafast isolation technology, combined two membrane filter configurations. Larger sEVs stayed in the central chamber while smaller particles and fluids were able to flow through the nonporous anodic aluminium oxide membrane due to periodic negative pressure oscillations caused by switching between periods of negative pressure and air pressure.[58] The low yields and membrane pore blockage issues with the conventional approaches are resolved by these techniques.
Magnetic beads immunization
The utilization of magnetic bead-based immunoaffinity enrichment has gained significant interest due to its notable advantages in terms of convenience and high efficiency. This method involves the use of magnetic beads coated with specific antibodies that target surface markers of sEVs. Through this immunomagnetic approach, the process of capturing and enriching sEVs becomes more efficient and effective. Through the application of magnetic beads that specifically bound to the CD63 protein on sEVs derived from serum of mice having breast cancer, a nanodevice was able to separate target sEVs. The target sEVs could be eluted from magnetic beads by controlling light excitation while preserving their integrity.[59] The problem of costly antibody applications on the beads is resolved by the aptamer approach. sEV-containing solutions can be selectively recognized and CD63 positive sEVs can be isolated using beads linked with CD63-1 aptamer.[60] In another approach, magnetic beads conjugated with a synthetic peptide, Vn96, were used to isolate sEVs from MCF7 cell culture medium. This method achieved high efficiency in isolating sEVs without affecting their morphology.[61] Numerous research endeavours have been dedicated to enhancing the functionality of magnetic beads. According to one study, immunomagnetic hedgehog particles (IMHPs) with nano-spikes can improve antibody-antigen based sEV binding and targeting by offering a greater surface area for immobilization of antibodies. These IMHPs demonstrated a capture efficiency of 91.7% in extracting sEVs from MCF-7 cells.[62] These advancements in magnetic bead-based immunomagnetic techniques contribute to the effectiveness and functionality of sEV isolation.
Electro-deposition
Electro-deposition is a technique that leverages electrostatic forces to isolate sEVs based on their electrical properties. These forces facilitate the movement of charged sEVs towards a conductive electrode, which can be modified to enhance their adhesion. By selectively depositing sEVs onto the conductive electrode, this method enables effective isolation and subsequent analysis and characterization of sEVs.[63] For the purpose of isolating superparamagnetic nanobeads, a superparamagnetic track-etched membrane has been created. These beads possess a high capture capacity, short incubation time, and achieve capture rates of up to 99%, making them a viable option for isolating sEVs from physiological samples.[64] Improved detection sensitivity was demonstrated by an electrode modified with chitosan composite, ionic liquid, and multi-walled carbon nanotubes. Breast cancer cell-derived sEVs containing HER-2 and EpCAM were found with high selectivity and sensitivity. Additionally, this approach presents the possibility of multiplex diagnosis detection of several sEV biomarkers.[65] Electro-deposition enables the precise isolation of sEVs while maintaining their integrity, minimizing any potential damage or alterations that may occur during the deposition process.
Acoustic-based isolation
Acoustic forces can be utilized for the separation of sEVs from biological samples as well. By generating acoustic waves, spatial pressure nodes and antinodes are created, effectively causing sEVs to migrate towards specific regions for isolation. Due to their smaller size and lower density compared to cells and debris, sEVs can be selectively separated from larger and denser particles. This enables the efficient isolation of sEVs using acoustic forces.[66] An example of this is the Acoustic Separation and Concentration of sEVs for Nucleotide Detection (ASCENDx) device, which utilizes a rotating microfluidic disc to enrich sEVs. In order to enable centrifugation and fluid actuation within the microfluidic channels on the disc surface, surface acoustic waves and the fluid layer on which the disc floats can be coupled to form the acoustofluidic disc rotation. With excellent selectivity and specificity of 95.8% and 100%, respectively, the enriched sEVs demonstrated diagnostic potential for identifying circulating colorectal cancer miRNA biomarkers from patient plasma samples.[67] From undiluted blood samples, sEVs can also be directly isolated using a different acoustofluidic technology. The platform consists of two sequential surface acoustic wave microfluidic modules: one for isolating sEVs and the other for removing cells. The sEV-isolation module purifies the sEVs by eliminating other EV subgroups, while the cell-removal module eliminates microscale blood components. This approach effectively yields high purity and quantity of sEVs from undiluted blood samples.[68]
New techniques for detecting sEV proteins
Conventional techniques for detecting sEV proteins involve a variety of laboratory methods, including western blotting and ELISA. However, these methods often have limitations such as limited sensitivity and difficulty in obtaining accurate and precise quantitative data.[69,70] To overcome these limitations, some new techniques and methodologies have been developed in recent years (Figure 3).
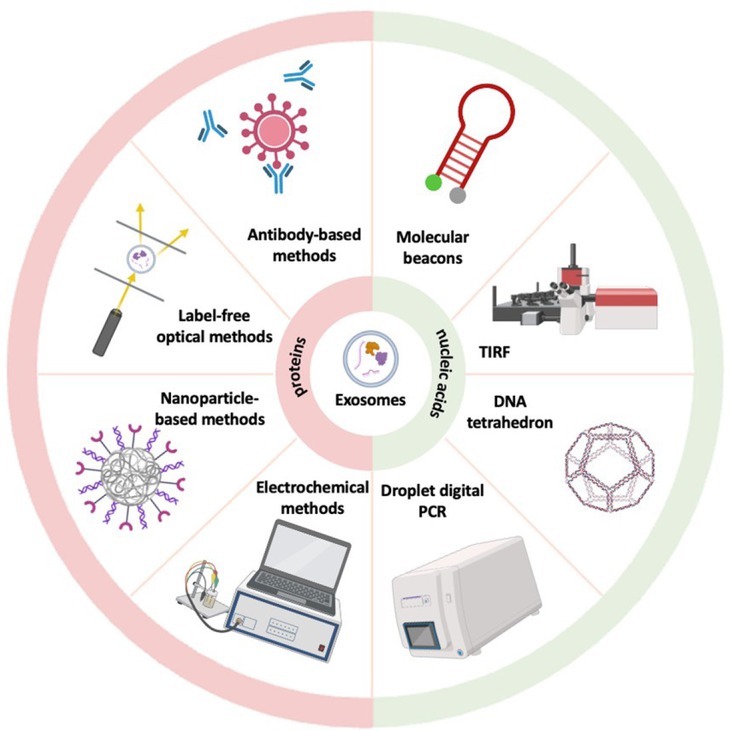
New methods for detection of sEV proteins and nucleic acids. The figure shows some new sEV proteins and nucleic acids detection methods, including antibody-based methods, label-free optical methods, nanoparticle-based methods, electrochemical methods for proteins, and droplet digital PCR, DNA tetrahedron, total internal reflection fluorescent and molecular beacons for nucleic acids. sEV: small extracellular vesicles.
Label-free optical methods
(SPR) is an optical sensing method that detects changes in the refractive index near a sensor surface caused by the binding of biomolecules, such as sEVs or their proteins.[71] This technique has been used to precisely and sensitively detect HER2-positive sEVs. This technique offers a potential breast cancer diagnostic approach by differentiating patients with HER2-positive breast cancer from healthy individuals.[72] This sensing approach can be expanded to accurately detect more sEV subtypes by simply altering the aptamer types. Utilizing sEV epidermal growth factor receptor and PD-L1 as biomarkers, SPR has also been used to diagnose lung cancer.[73] Another analytical method for the identification and characterization of molecules is surface-enhanced Raman scattering (SERS). SERS uses chemical and electromagnetic processes to increase the Raman signal of tiny molecules affixed to the uneven metal surface.[74] It has been used extensively in the search for sEV biomarkers. Using this method, a portable Raman sEV assay for sEV detection and protein profiling was developed. SERS can be used to diagnose breast cancer, and biomarkers for HER2 and EpCAM have been found to have diagnostic potential on sEVs in plasma from patients with HER2-positive breast cancer. According to these proof-of-concept investigations, this assay could accelerate research on sEVs and open the door for the creation of innovative liquid biopsies for cancer monitoring and detection.[75] It is possible to precisely identify sEVs generated from cancer cells by combining statistical pattern analysis with SERS. Through principal component analysis of the entire SERS spectra of the sEVs, lung cancer cell-derived sEVs were identified with 95.3% sensitivity and 97.3% specificity from sEVs originated from normal cells.[76]
Antibody-based methods
The unique binding of antibodies to sEV surface indicators is the basis for antibody-based techniques such as fluorescence detection and lateral flow assays, which allows the detection and study of these markers in sEVs. The presence of target analyte is determined by antibody-antigen interactions in lateral flow tests. This technique has been used to detect isolated sEVs from a malignant melanoma cell line, with a detection limit of 8.54x10^5 sEVs/μL.[77] Additionally, it has been used to create a point-of-care platform for the detection and tracking of colorectal cancer. Together with lateral flow experiments, CD147-containing sEVs were employed as a biomarker to identify and monitor colorectal cancer. This point-of-care tool was used to quantify the CD147 antigen embedded in sEVs that were isolated from plasma.[78] By using fluorescent labelling, sEVs can be visualized using the fluorescent detection method. This method has been utilized to detect sEVs from biological samples. The estimated limit of detection for sEVs using this method was 1.29 × 103 particles/μL. Additionally, the difference in sEV concentration between sera of healthy individuals and cancer patients was evaluated.[79] This technique could potentially be developed into a platform for the precise and specific identification of sEVs in biological samples for the diagnosis of cancer. A study showed that the fluorescence approach may reliably identify plasma sEVs, with an AUC of 0.85 for cancer diagnosis, to differentiate lung cancer patients from healthy persons.[80] By employing other tumour-related sEV proteins as recognition targets, it is possible to isolate and identify sEVs from specific subpopulations derived from tumour cells, thereby improving the sensitivity and specificity of tumour diagnosis.
Nanoparticle-based methods
Colorimetric assays can be employed to detect sEVs using nanozymes, which are nanomaterials possessing inherent catalytic activity similar to those of enzymes. Through surface modification with certain ligands, the nanozyme can selectively bind to sEV surface indicators. In a study, the CD63 aptamer was utilized to identify sEVs. The hybrid nanozyme’s peroxidase activity was increased and a colorimetric signal was produced with the aid of the CD63 aptamer-bound sEVs. As a result, sEVs with a detection limit of 3.37 × 103 particles/μL could be found.[81] Similarly, EpCAM aptamer was utilized in the nanozyme-based colorimetric assay to provide specific detection, hence enabling the differentiation of breast cancer patients from healthy individuals.[82] Quantum dots (QDs), one of the several varieties of nanoparticle-based optical labels, are advantageous for sEV detection because of their small diameter (2–10 nm), which enables effective sEV labelling and detection in a smaller size range.[83] Different surface protein markers on sEVs from various breast cancer cell lines were specifically and quantitatively detected using the QD-based technique, and cancer-associated surface protein indicators can be used to distinguish sEVs produced from cancer cells from normal sEVs. By employing QDs to analyse HER2 expression on plasma sEVs, HER2-positive breast cancer was identified. Patients with the disease had HER2 expression that was around five times higher than that of healthy donors, with an AUC value of 0.96875.[84]
Electrochemical methods
Electrochemical Impedance Spectroscopy (EIS) is a method that assesses the impedance response of a system when subjected to an alternating current signal across various frequencies. This technique proves valuable in detecting impedance changes arising from the interactions between sEVs and electrode surfaces. Considering that sEVs possess distinct compositions, particularly in terms of their membrane and cytosolic charge-dependent contents, variations in their opacity can serve as distinguishing factors. Consequently, EIS enables the differentiation of sEVs derived from different cellular origins based on their unique characteristics.[85] Electrochemical biosensors, on the other hand, utilize electrochemical reactions taking place at the electrode surface, which can be influenced by the presence of sEVs. In a recent study, tumour cell-derived sEVs were successfully detected using a combination of cyclic nicking enzyme cleavage and a hybridization chain reaction for dual-signal amplification. For this assay, a hairpin aptamer probe (HAP) containing an aptamer was designed. The aptamer specifically binds to PTK7, a protein found on the surface of sEVs, causing a conformational change in the HAP. This conformational change enables hybridization between the HAP and the linker DNA, initiating cyclic cleavage of the nicking endonuclease on the linker DNA. Consequently, sEV detection is transformed into DNA detection. By incorporating this approach with HCR signal amplification, the study achieved highly sensitive electrochemical detection of CCRF-CEM sEVs, with a limit of detection as low as 1.1 × 104 particles/mL.[86]
New techniques for analysing sEV nucleic acids
SEVs contain not only protein cargoes, but also nucleic acids, which have shown promising result as a specific biomarker for cancer diagnosis and prognosis prediction. To quantify the expression levels of sEV nucleic acids, techniques such as qRT-PCR, microarray, and next-generation sequencing have been widely used.[87] However, qRT-PCR is limited to detecting nucleic acids with known sequences, while NGS is costly and involves complex library construction.[88] Microarrays, although capable of analysing thousands of nucleic acids simultaneously, have low sensitivity.[89] To address these limitations, efforts are underway to develop highly sensitive and convenient methods for sEV nucleic acid detection that overcome these challenges (Figure 3).
Molecular beacons
Molecular beacons are hairpin-shaped nucleic acid probes that detect specific target sequences. They form a stem-loop structure with a fluorophore and quencher in close proximity. When the target binds to the probes, the stem opens, resulting in the activation of fluorescence.[90] It has been shown that miRNA-targeting molecular beacons can detect several miRNAs simultaneously in sEVs. In sEVs generated from MCF-7, molecular beacons hybridized with numerous miRNAs despite the high concentration of human serum.[91] Even in the presence of human urine, molecular beacons were able to identify the markers miRNA-375 and miRNA-574-3p, which are present in sEVs produced by prostate cancer cells. This implies that they can be used to do liquid biopsies for prostate cancer using human urine.[92]
Solid-state nanopore sensing
Solid-state nanopore sensing involves applying a voltage across a solid-state material that contains a nanopore. When nucleic acid passes through the nanopore, it temporarily blocks the flow of ions or electrons, causing a detectable change in the electrical current. This change in current can be measured and analyzed.[93] This technique has been utilized to detect the only two cysteine-containing peptides from LRG-1, an emerging protein biomarker, that are uniquely present in the urine of ovarian cancer patients. The technique provided improved selectivity for detecting biomarkers in ovarian cancer.[94] Moreover, solid-state nanopores have been explored as single-molecule counters for future digital diagnostic technologies, as evidenced by this technology’s capacity to quantify more than six distinct microRNA concentrations.[95]
Droplet digital PCR
In digital droplet PCR (ddPCR), the nucleic acid sample of interest is divided into numerous separate reaction droplets, each containing a few target molecules or none at all. These droplets undergo PCR amplification within a thermal cycler, and upon completion, fluorescence-based detection methods are employed to analyze the results. Each individual droplet is examined to determine whether it exhibits a fluorescence signal or not. By quantifying the number of positive and negative droplets, the absolute quantity of target molecules in the initial sample can be accurately calculated using Poisson statistics.[96] A study used the ddPCR technology with sEV DNA to develop a sensitive and accurate approach for diagnosing tuberculosis. sEV and total DNA that was isolated from respiratory samples were targeted to the IS6110 region. The use of sEV DNA in ddPCR resulted in greater sensitivity and specificity of 98.0% and 76.9%, respectively, similar to whole DNA.[97] This suggests that the combination of ddPCR platform with sEV DNA has the potential to provide a sensitive and accurate methodology for diagnosis of cancer. Another study evaluated the efficacy of various methods for detecting EGFR mutation in pleural fluid and plasma samples; the results showed that ddPCR in conjunction with sEV DNA had the highest sensitivity. With NSCLC patients, this method may be able to detect genetic alterations linked to resistance to EGFR inhibitor treatment.[98]
DNA tetrahedron
DNA tetrahedron is a versatile DNA nanostructure that offers precise control over its architecture. Through chemical modifications and DNA self-assembly, it can be engineered to provide a wide range of amplified signal tags.[99] A study used DNA tetrahedron nanoprobe (DTNP) based on fluorescence resonance energy transfer (FRET) to establish a sensitive detection approach for has-miR-146b-5p, a tumour-related microRNA. The target miRNA was intended to cause a structural alteration in the DTNP, leading to a significant enhancement in the FRET signal. This method facilitated the assessment of miRNA expression levels in various cell lines and demonstrated a low limit of detection.[100]
Total internal reflection fluorescence
Total internal reflection fluorescence (TIRF) imaging assay relies on the principle of total internal reflection, where a laser beam is directed at a specific angle onto a glass or prism surface, creating an evanescent wave that penetrated only a short distance into the sample.[101] With the use of this method, single sEVs and their miRNA contents in serum microsamples can be directly visualized and measured. Serum sEV miR-21 is a commonly used cancer biomarker. The TIRF imaging test was used to analyse miR-21 and demonstrated a better performance than traditional real-time PCR assays in differentiating cancer patients from healthy individuals.[102]
Microfluidic devices utilizing sEVs for cancer diagnosis
Microfluidic devices have been developed to integrate the isolation and analysis of sEVs into a platform where sEVs can be directly characterized and analyzed for downstream genomics and proteomics. Microfluidic devices can enrich sEVs from low-abundance sample for enhancing the sensitivity of downstream analysis. Tumour-derived sEVs can be directly quantified from as little as 1 μL of plasma using an approach known as nanoplasmon-enhanced scattering (nPES), which also uses antibody-conjugated gold nanospheres and nanorods to capture sEVs. By detecting ephrin type-A receptor 2, this assay can differentiate between patients with pancreatitis, healthy individuals, and those with pancreatic cancer.[103] Some microfluidic devices enable the identification and quantification of specific cancer-associated biomarkers carried by sEVs. The mRNA levels of MGMT (O6-methylguanine DNA methyltransferase) and APNG (alkylpurine-DNA-N-glycosylase), whose levels in tissue are inversely related to the effectiveness of drug treatment in glioblastoma multiforme, were examined in enriched tumour sEVs derived from blood using a microfluidic chip.[104] By examining the expression and proteolytic activity of MMP14 on sEVs using three-dimensional nanopatterned devices, tumour growth and metastasis may also be tracked.[105]
Microfluidic devices offer significant advantages for the rapid and portable cancer diagnostics using sEVs, making them suitable for point-of-care applications. These devices hold great potential for early cancer detection and monitoring treatment response. A nanoparticle-based biochip enables the capture of circulating sEV and enhances the fluorescence signals of encapsulated RNAs. This is achieved by a catalysed hairpin DNA circuit confined within cationic lipid-polymer hybrid nanoparticles tethered on the chip which amplifies these signals in situ, all in a single step. This biochip can selectively and sensitively identify low expression of glypican-1 mRNA in serum sEVs, allowing for the discrimination of patients with early- and late-stage pancreatic cancer from healthy individuals and patients with benign pancreatic disease.[106] Another microfluidic device, using self-assembled three-dimensional herringbone nanopatterns, can identify low concentrations of tumour-associated sEVs in plasma. This device suggests sEV folate receptor alpha as a potential biomarker for early detection and progression monitoring of ovarian cancer.[34] Microfluidic devices provide an efficient platform for utilizing sEVs as cancer biomarkers. Their integration with isolation, enrichment, and detection techniques enables sensitive and specific analysis of sEV cargo, facilitating the development of non-invasive cancer diagnostic approaches.
Conclusion
sEV biomarkers have emerged as valuable indicators of cancer progression and treatment response. The utilization of sEVs in liquid biopsy enables non-invasive monitoring of disease and holds promise for personalized treatment strategies. Moreover, the translation of sEVs into clinical trials underscores their potential clinical utility. These trials aim to validate the diagnostic and prognostic value of sEV biomarkers, as well as explore their therapeutic potential in drug delivery and immunotherapy. Significant advancements have been made in sEV isolation and enrichment techniques, and in the detection of sEV proteins and nucleic acids. These advancements have improved the sensitivity and specificity of sEV-based diagnostics, allowing for more accurate and reliable detection and analysis of cancer-related biomarkers. Microfluidic chips utilizing sEVs have shown promise for cancer diagnosis, offering rapid and efficient analysis. These miniaturized devices integrate multiple functions into a single platform, enabling streamlined and high-throughput analysis of sEV biomarkers. However, further research and development are crucial to fully harness the potential of sEV biomarkers in clinical applications. Standardized protocols for sEV isolation, characterization, and analysis need to be established to ensure reproducibility and comparability across different studies and clinical settings. Additionally, long-term studies are required to evaluate the clinical outcomes and cost-effectiveness of integrating sEV biomarkers into routine clinical practice. In conclusion, sEV biomarkers have paved the way for a new era in cancer management.
Funding statement: The work was funded by Material Innovation Institute for Life Sciences and Energy (MILES) Fellowship Program and National Natural Science Foundation of China General Program (Project No. 82072626).
Acknowledgements
Figures 1, 2 and 3 in this article were created with Biorender.com.
-
Author Contributions
Conceptualization: CWLT, JWPY; Writing - original draft: CWLT; Writing - review and editing: CWLT, JWPY; Supervision and funding acquisition: JWPY.
-
Ethical Approval
Not applicable.
-
Informed Consent
Not applicable.
-
Conflict of Interest
The authors declare no conflicts of interest.
-
Use of Large Language Models, AI and Machine Learning Tools
None declared.
-
Data Availability Statement
Data sharing not applicable – no new data generated.
References
1 Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci 2013;14:14240–14269.10.3390/ijms140714240Search in Google Scholar PubMed PubMed Central
2 Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750.10.1080/20013078.2018.1461450Search in Google Scholar
3 Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 2013;126:5553–5565.10.1242/jcs.128868Search in Google Scholar PubMed
4 Tschuschke M, Kocherova I, Bryja A, Mozdziak P, Angelova Volponi A, Janowicz K, et al. Methods of Isolation and Clinical Application of Human Cellular Exosomes. J Clin Med 2020;9:436.10.3390/jcm9020436Search in Google Scholar PubMed PubMed Central
5 Duréndez-Sáez E, Calabuig-Fariñas S, Torres-Martínez S, Moreno-Manuel A, Herreros-Pomares A, Escorihuela E, et al. Analysis of exosomal cargo provides accurate clinical, histologic and mutational information in non-small cell lung cancer. Cancers (Basel) 2022;14:3216.10.3390/cancers14133216Search in Google Scholar PubMed PubMed Central
6 Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869–3875.10.1074/jbc.C113.532267Search in Google Scholar PubMed PubMed Central
7 Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–659.10.1038/ncb1596Search in Google Scholar PubMed
8 Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2011;2:180.10.1038/ncomms1180Search in Google Scholar PubMed PubMed Central
9 Dixson AC, Dawson TR, Di Vizio D, Weaver AM. Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nat Rev Mol Cell Biol 2023;24:454–476.10.1038/s41580-023-00576-0Search in Google Scholar PubMed PubMed Central
10 Li SP, Lin ZX, Jiang XY, Yu XY. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol Sin 2018;39:542–551.10.1038/aps.2017.178Search in Google Scholar PubMed PubMed Central
11 He X, Yang J, Ji M, Chen Y, Chen Y, Li H, Wang H. A potential delivery system based on cholera toxin: A macromolecule carrier with multiple activities. J Control Release 2022;343:551–563.10.1016/j.jconrel.2022.01.050Search in Google Scholar PubMed
12 Zhang C, Luo Y, Cao J, Wang X, Miao Z, Shao G. Exosomal lncRNA FAM225A accelerates esophageal squamous cell carcinoma progression and angiogenesis via sponging miR-206 to upregulate NETO2 and FOXP1 expression. Cancer Med 2020;9:8600–8611.10.1002/cam4.3463Search in Google Scholar PubMed PubMed Central
13 Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012;151:1542–1556.10.1016/j.cell.2012.11.024Search in Google Scholar PubMed
14 Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, et al. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res 2018;28:862–864.10.1038/s41422-018-0060-4Search in Google Scholar PubMed PubMed Central
15 Wen SW, Lima LG, Lobb RJ, Norris EL, Hastie ML, Krumeich S, et al. Breast cancer-derived exosomes reflect the cell-of-origin phenotype. Proteomics 2019;19:e1800180.10.1002/pmic.201800180Search in Google Scholar PubMed
16 Sonkin D, Thomas A, Teicher BA. Cancer treatments: Past, present, and future. Cancer Genet 2024:286–287:18–24.10.1016/j.cancergen.2024.06.002Search in Google Scholar PubMed PubMed Central
17 Raez LE, Brice K, Dumais K, Lopez-Cohen A, Wietecha D, Izquierdo PA, et al. Liquid biopsy versus tissue biopsy to determine front line therapy in metastatic non-small cell lung cancer (NSCLC). Clin Lung Cancer 2023;24:120–129.10.1016/j.cllc.2022.11.007Search in Google Scholar PubMed
18 Ahn JC, Teng PC, Chen PJ, Posadas E, Tseng HR, Lu SC, et al. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology 2021;73:422–436.10.1002/hep.31165Search in Google Scholar PubMed PubMed Central
19 Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–930.10.1126/science.aar3247Search in Google Scholar PubMed PubMed Central
20 Logozzi M, Angelini DF, Giuliani A, Mizzoni D, Di Raimo R, Maggi M, et al. Increased plasmatic levels of PSA-expressing exosomes distinguish prostate cancer patients from benign prostatic hyperplasia: A prospective study. Cancers (Basel) 2019;11:1449.10.3390/cancers11101449Search in Google Scholar PubMed PubMed Central
21 Thakur K, Singh MS, Feldstein-Davydova S, Hannes V, Hershkovitz D, Tsuriel S. Extracellular vesicle-derived DNA vs. CfDNA as a biomarker for the detection of colon cancer. Genes (Basel) 2021;12:1171.10.3390/genes12081171Search in Google Scholar PubMed PubMed Central
22 Moldovan N, Verkuijlen S, van der Pol Y, Bosch L, van Weering JRT, Bahce I, et al. Comparison of cell-free and small extracellular-vesicle-associated DNA by sequencing plasma of lung cancer patients. iScience 2024;27:110742.10.1016/j.isci.2024.110742Search in Google Scholar PubMed PubMed Central
23 Zhou H, Yuen PS, Pisitkun T, Gonzales PA, Yasuda H, Dear JW, et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int 2006;69:1471–1476.10.1038/sj.ki.5000273Search in Google Scholar PubMed PubMed Central
24 Hsu CC, Wu Y. Recent advances in nanotechnology-enabled biosensors for detection of exosomes as new cancer liquid biopsy. Exp Biol Med (Maywood) 2022;247:2152–2172.10.1177/15353702221110813Search in Google Scholar PubMed PubMed Central
25 Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015;523:177–182.10.1038/nature14581Search in Google Scholar PubMed PubMed Central
26 Li J, Chen Y, Guo X, Zhou L, Jia Z, Peng Z, et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med 2017;21:838–847.10.1111/jcmm.12941Search in Google Scholar PubMed PubMed Central
27 Xiao D, Dong Z, Zhen L, Xia G, Huang X, Wang T, et al. Combined exosomal GPC1, CD82, and Serum CA19-9 as multiplex targets: A specific, sensitive, and reproducible detection panel for the diagnosis of pancreatic cancer. Mol Cancer Res 2020;18:300–310.10.1158/1541-7786.MCR-19-0588Search in Google Scholar PubMed
28 Ouyang H, Gore J, Deitz S, Korc M. microRNA- 10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-β actions. Oncogene 2014;33:4664–4674.10.1038/onc.2013.405Search in Google Scholar PubMed PubMed Central
29 Pang W, Su J, Wang Y, Feng H, Dai X, Yuan Y, et al. Pancreatic cancer-secreted miR- 155 implicates in the conversion from normal fibroblasts to cancer-associated fibroblasts. Cancer Sci 2015;106:1362–1369.10.1111/cas.12747Search in Google Scholar PubMed PubMed Central
30 Wu M, Tan X, Liu P, Yang Y, Huang Y, Liu X, et al. Role of exosomal microRNA-125b-5p in conferring the metastatic phenotype among pancreatic cancer cells with different potential of metastasis. Life Sci 2020;255:117857.10.1016/j.lfs.2020.117857Search in Google Scholar PubMed
31 Gao S, Sun Y, Liu X, Zhang D, Yang X. EpCAM and COX 2 expression are positively correlated in human breast cancer. Mol Med Rep 2017;15:3755–3760.10.3892/mmr.2017.6447Search in Google Scholar PubMed
32 Runz S, Keller S, Rupp C, Stoeck A, Issa Y, Koensgen D, et al. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol Oncol 2007;107:563–571.10.1016/j.ygyno.2007.08.064Search in Google Scholar PubMed
33 Gan J, Zeng X, Wang X, Wu Y, Lei P, Wang Z, et al. Effective diagnosis of prostate cancer based on mRNAs from urinary exosomes. Front Med (Lausanne) 2022;9:736110.10.3389/fmed.2022.736110Search in Google Scholar PubMed PubMed Central
34 Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK, et al. Ultra-sensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat Biomed Eng 2019;3:438–451.10.1038/s41551-019-0356-9Search in Google Scholar PubMed PubMed Central
35 Tian F, Zhang S, Liu C, Han Z, Liu Y, Deng J, et al. Protein analysis of extracellular vesicles to monitor and predict therapeutic response in metastatic breast cancer. Nat Commun 2021;12:2536.10.1038/s41467-021-22913-7Search in Google Scholar PubMed PubMed Central
36 Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem 2018;119:4711–4716.10.1002/jcb.26650Search in Google Scholar PubMed
37 de Miguel-Perez D, Russo A, Arrieta O, Ak M, Barron F, Gunasekaran M, et al. Extracellular vesicle PD-L1 dynamics predict durable response to immune-checkpoint inhibitors and survival in patients with non-small cell lung cancer. J Exp Clin Cancer Res 2022;41:186.10.1186/s13046-022-02379-1Search in Google Scholar PubMed PubMed Central
38 Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer 2015;112:532–538.10.1038/bjc.2014.621Search in Google Scholar PubMed PubMed Central
39 Zhang Y, Liu H, Liu X, Guo Y, Wang Y, Dai Y, et al. Identification of an exosomal long non-coding RNAs panel for predicting recurrence risk in patients with colorectal cancer. Aging (Albany NY) 2020;12:6067–6088.10.18632/aging.103006Search in Google Scholar PubMed PubMed Central
40 Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med 2019;23:1396–1405.10.1111/jcmm.14042Search in Google Scholar PubMed PubMed Central
41 Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang X, et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene 2018;37:3822–3838.10.1038/s41388-018-0237-9Search in Google Scholar PubMed
42 Yen EY, Miaw SC, Yu JS, Lai IR. Exosomal TGF-beta1 is correlated with lymphatic metastasis of gastric cancers. Am J Cancer Res 2017;7:2199–2208.Search in Google Scholar
43 Chu J, Yang D, Wang L, Xia J. Nomograms predicting survival for all four subtypes of breast cancer: a SEER-based population study. Ann Transl Med 2020;8:544.10.21037/atm-20-2808Search in Google Scholar PubMed PubMed Central
44 Stevic I, Müller V, Weber K, Fasching PA, Karn T, Marmé F, et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med 2018;16:179.10.1186/s12916-018-1163-ySearch in Google Scholar PubMed PubMed Central
45 Yang G, Tan J, Guo J, Wu Z, Zhan Q. Exosome-mediated transfer of circ_0063526 enhances cisplatin resistance in gastric cancer cells via regulating miR-449a/SHMT2 axis. Anticancer Drugs 2022;33:1047–1057.10.1097/CAD.0000000000001386Search in Google Scholar PubMed
46 Yuwen D, Ma Y, Wang D, Gao J, Li X, Xue W, et al. Prognostic role of circulating exosomal miR-425-3p for the response of NSCLC to platinum-based chemotherapy. Cancer Epidemiol Biomarkers Prev 2019;28:163–173.10.1158/1055-9965.EPI-18-0569Search in Google Scholar PubMed
47 Zheng Q, Ding H, Wang L, Yan Y, Wan Y, Yi Y, et al. Circulating exosomal miR-96 as a novel biomarker for radioresistant non-small-cell lung cancer. J Oncol 2021;2021:5893981.10.1155/2021/5893981Search in Google Scholar PubMed PubMed Central
48 Yang Q, Chen M, Gu J, Niu K, Zhao X, Zheng L, et al. Novel Biomarkers of dynamic blood PD-L1 expression for immune checkpoint inhibitors in advanced non-small-cell lung cancer patients. Front Immunol 2021;12:665133.10.3389/fimmu.2021.665133Search in Google Scholar PubMed PubMed Central
49 Zhu J, Tang B, Gao Y, Xu S, Tu J, Wang Y, et al. Predictive Models for HCC prognosis, recurrence risk, and immune infiltration based on two exosomal genes; MYL6B and THOC2. J Inflamm Res 2021;14:4089–4109.10.2147/JIR.S315957Search in Google Scholar PubMed PubMed Central
50 Kaya C, Dorsaint P, Mercurio S, Campbell AM, Eng KW, Nikiforova MN, Elemento O, Nikiforov YE, Sboner A. Limitations of Detecting Genetic Variants from the RNA Sequencing Data in Tissue and Fine-Needle Aspiration Samples. Thyroid 2021;31:589–595.10.1089/thy.2020.0307Search in Google Scholar PubMed PubMed Central
51 Lohmann K, Klein C. Next generation sequencing and the future of genetic diagnosis. Neurotherapeutics 2014;11:699–707.10.1007/s13311-014-0288-8Search in Google Scholar PubMed PubMed Central
52 Yakubovich EI, Polischouk AG, Evtushenko VI. Principles and problems of exosome isolation from biological fluids. Biochem (Mosc) Suppl Ser A Membr Cell Biol 2022;16:115–126.10.1134/S1990747822030096Search in Google Scholar PubMed PubMed Central
53 Yang Y, Wang Y, Wei S, Zhou C, Yu J, Wang G, et al. Extracellular vesicles isolated by size-exclusion chromatography present suitability for RNomics analysis in plasma. J Transl Med 2021;19:104.10.1186/s12967-021-02775-9Search in Google Scholar PubMed PubMed Central
54 Tangwattanachuleeporn M, Muanwien P, Teethaisong Y, Somparn P. Optimizing concentration of polyethelene glycol for exosome isolation from plasma for Downstream application. Medicina (Kaunas) 2022:58:1600.10.3390/medicina58111600Search in Google Scholar PubMed PubMed Central
55 Hochstetter A, Vernekar R, Austin RH, Becker H, Beech JP, Fedosov DA, et al. Deterministic lateral displacement: Challenges and perspectives. ACS Nano 2020;14:10784–10795.10.1021/acsnano.0c05186Search in Google Scholar PubMed
56 Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, et al. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat Nanotechnol 2016;11:936–940.10.1038/nnano.2016.134Search in Google Scholar PubMed
57 Smith JT, Wunsch BH, Dogra N, Ahsen ME, Lee K, Yadav KK, et al. Integrated nanoscale deterministic lateral displacement arrays for separation of extracellular vesicles from clinically-relevant volumes of biological samples. Lab Chip 2018;18:3913–3925.10.1039/C8LC01017JSearch in Google Scholar PubMed
58 Chen Y, Zhu Q, Cheng L, Wang Y, Li M, Yang Q, et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat Methods 2021;18:212– 218.10.1038/s41592-020-01034-xSearch in Google Scholar PubMed
59 Wang C, Zhang D, Yang H, Shi L, Li L, Yu C, et al. A light-activated magnetic bead strategy utilized in spatio-temporal controllable exosomes isolation. Front Bioeng Biotechnol 2022;10:1006374.10.3389/fbioe.2022.1006374Search in Google Scholar PubMed PubMed Central
60 Song Z, Mao J, Barrero RA, Wang P, Zhang F, Wang T. Development of a CD63 aptamer for efficient cancer immunochemistry and immunoaffinity-based exosome isolation. Molecules 2020;25:5585.10.3390/molecules25235585Search in Google Scholar PubMed PubMed Central
61 Bathini S, Pakkiriswami S, Ouellette RJ, Ghosh A, Packirisamy M. Magnetic particle based liquid biopsy chip for isolation of extracellular vesicles and characterization by gene amplification. Biosens Bioelectron 2021;194:113585.10.1016/j.bios.2021.113585Search in Google Scholar PubMed
62 Cheng J, Zhu N, Zhang Y, Yu Y, Kang K, Yi Q, et al. Hedgehog-inspired immunomagnetic beads for high-efficient capture and release of exosomes. J Mater Chem B 2022;10:4059–4069.10.1039/D2TB00226DSearch in Google Scholar PubMed
63 Tayebi M, Yang D, Collins DJ, Ai Y. Deterministic sorting of submicrometer particles and extracellular vesicles using a combined electric and acoustic field. Nano Lett 2021;21:6835–6842.10.1021/acs.nanolett.1c01827Search in Google Scholar PubMed
64 Zhang C, Huo X, Zhu Y, Higginbotham JN, Cao Z, Lu X, et al. Electrodeposited magnetic nanoporous membrane for high-yield and high-throughput immunocapture of extracellular vesicles and lipoproteins. Commun Biol 2022;5:1358.10.1038/s42003-022-04321-9Search in Google Scholar PubMed PubMed Central
65 Hashkavayi AB, Cha BS, Lee ES, Park KS. Dual rolling circle amplification-enabled ultrasensitive multiplex detection of exosome biomarkers using electrochemical aptasensors. Anal Chim Acta 2022;1205:339762.10.1016/j.aca.2022.339762Search in Google Scholar PubMed
66 Ku A, Lim HC, Evander M, Lilja H, Laurell T, Scheding S, et al. Acoustic enrichment of extracellular vesicles from biological fluids. Anal Chem 2018;90:8011–8019.10.1021/acs.analchem.8b00914Search in Google Scholar PubMed PubMed Central
67 Naquin TD, Canning AJ, Gu Y, Chen J, Naquin CM, Xia J, et al. Acoustic separation and concentration of exosomes for nucleotide detection; ASCENDx. Sci Adv 2024;10:eadm8597.10.1126/sciadv.adm8597Search in Google Scholar PubMed PubMed Central
68 Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci USA 2017;114:10584–10589.10.1073/pnas.1709210114Search in Google Scholar PubMed PubMed Central
69 Ghosh R, Gilda JE, Gomes AV. The necessity of and strategies for improving confidence in the accuracy of western blots. Expert Rev Proteomics 2014;11:549–560.10.1586/14789450.2014.939635Search in Google Scholar PubMed PubMed Central
70 Güven E, Duus K, Lydolph MC, Jørgensen CS, Laursen I, Houen G. Non-specific binding in solid phase immunoassays for autoantibodies correlates with inflammation markers. J Immunol Methods 2014;403:26–36.10.1016/j.jim.2013.11.014Search in Google Scholar PubMed
71 Nguyen HH, Park J, Kang S, Kim M. Surface plasmon resonance: a versatile technique for biosensor applications. Sensors (Basel) 2015;15:10481–10510.10.3390/s150510481Search in Google Scholar PubMed PubMed Central
72 Chen W, Li Z, Cheng W, Wu T, Li J, Li X, et al. Surface plasmon resonance biosensor for exosome detection based on reformative tyramine signal amplification activated by molecular aptamer beacon. J Nanobiotechnology 2021;19:450.10.1186/s12951-021-01210-xSearch in Google Scholar PubMed PubMed Central
73 Wu Y, Zeng X, Gan Q. A Compact Surface Plasmon Resonance Biosensor for Sensitive Detection of Exosomal Proteins for Cancer Diagnosis. Methods Mol Biol 2022;2393:3–14.10.1007/978-1-0716-1803-5_1Search in Google Scholar PubMed
74 Li J, Li Y, Li P, Zhang Y, Du L, Wang Y, et al. Exosome detection via surface-enhanced Raman spectroscopy for cancer diagnosis. Acta Biomater 2022;144:1–14.10.1016/j.actbio.2022.03.036Search in Google Scholar PubMed
75 Kwizera EA, O'Connor R, Vinduska V, Williams M, Butch ER, Snyder SE, et al. Molecular detection and analysis of exosomes using surface-enhanced raman scattering gold nanorods and a miniaturized device. Theranostics 2018;8:2722–2738.10.7150/thno.21358Search in Google Scholar PubMed PubMed Central
76 Park J, Hwang M, Choi B, Jeong H, Jung JH, Kim HK, et al. Exosome classification by pattern analysis of surface-enhanced raman spectroscopy data for lung cancer diagnosis. Anal Chem 2017;89:6695–6701.10.1021/acs.analchem.7b00911Search in Google Scholar PubMed
77 Oliveira-Rodríguez M, López-Cobo S, Reyburn HT, Costa-García A, López-Martín S, Yáñez-Mó M, et al. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J Extracell Vesicles 2016;5:31803.10.3402/jev.v5.31803Search in Google Scholar PubMed PubMed Central
78 Moyano A, Serrano-Pertierra E, Duque JM, Ramos V, Teruel-Barandiarán E, Fernández-Sánchez MT, et al. Magnetic lateral flow immunoassay for small extracellular vesicles quantification: Application to colorectal cancer biomarker detection. Sensors (Basel) 2021;21:3756.10.3390/s21113756Search in Google Scholar PubMed PubMed Central
79 Wang X, Shang H, Ma C, Chen L. A Fluorescence assay for exosome detection based on bivalent cholesterol anchor triggered target conversion and enzyme-free signal amplification. Anal Chem 2021;93:8493–8500.10.1021/acs.analchem.1c00796Search in Google Scholar PubMed
80 Wu Y, Gao Z, Chai Y, Zhang A, He S, Liu X, et al. One-step and label-free ratiometric fluorescence assay for the detection of plasma exosome towards cancer diagnosis. Talanta 2024;271:125700.10.1016/j.talanta.2024.125700Search in Google Scholar PubMed
81 Li C, Guo Z, Pu S, Zhou C, Cheng X, Zhao R, et al. Molybdenum disulfide-integrated iron organic framework hybrid nanozyme-based aptasensor for colorimetric detection of exosomes. Biosensors (Basel) 2023;13:800.10.3390/bios13080800Search in Google Scholar PubMed PubMed Central
82 Zhang X, Zhu X, Li Y, Hai X, Bi S. A colorimetric and photothermal dual-mode biosensing platform based on nanozyme-functionalized flower-like DNA structures for tumor-derived exosome detection. Talanta 2023;258:124456.10.1016/j.talanta.2023.124456Search in Google Scholar PubMed
83 Kairdolf BA, Smith AM, Stokes TH, Wang MD, Young AN, Nie S. Semiconductor quantum dots for bioimaging and biodiagnostic applications. Annu Rev Anal Chem (Palo Alto Calif) 2013;6:143–162.10.1146/annurev-anchem-060908-155136Search in Google Scholar PubMed PubMed Central
84 Vinduska V, Gallops CE, O'Connor R, Wang Y, Huang X. Exosomal surface protein detection with quantum dots and immunomagnetic capture for cancer detection. Nanomaterials (Basel) 2021;11:1853.10.3390/nano11071853Search in Google Scholar PubMed PubMed Central
85 Shi L, Esfandiari L. A label-free and low-power microelectronic impedance spectroscopy for characterization of exosomes. PLoS One 2022;17:e0270844.10.1371/journal.pone.0270844Search in Google Scholar PubMed PubMed Central
86 Sun D, Guo Q, Zhang H, Cai C. Electrochemical detection of tumor cell-derived exosomes based on cyclic enzyme scission and hybridization chain reaction dual-signal amplification. Chemosednsors 2023;11:415.10.3390/chemosensors11070415Search in Google Scholar
87 Onukwugha NE, Kang YT, Nagrath S. Emerging micro-nanotechnologies for extracellular vesicles in immuno-oncology: from target specific isolations to immunomodulation. Lab Chip 2022;22:3314–3339.10.1039/D2LC00232ASearch in Google Scholar PubMed PubMed Central
88 Head SR, Komori HK, LaMere SA, Whisenant T, Van Nieuwerburgh F, Salomon DR, et al. Library construction for next-generation sequencing: overviews and challenges. Biotechniques 2014;56:61–64, 66, 68, passim.10.2144/000114133Search in Google Scholar PubMed PubMed Central
89 Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet 2006;22:101– 109.10.1016/j.tig.2005.12.005Search in Google Scholar PubMed PubMed Central
90 Rhee WJ, Jeong S. Extracellular vesicle miRNA detection using molecular beacons. Methods Mol Biol 2017;1660:287–294.10.1007/978-1-4939-7253-1_23Search in Google Scholar PubMed
91 Lee JH, Kim JA, Jeong S, Rhee WJ. Simultaneous and multiplexed detection of exosome microRNAs using molecular beacons. Biosens Bioelectron 2016;86:202–210.10.1016/j.bios.2016.06.058Search in Google Scholar PubMed
92 Lee J, Kwon MH, Kim JA, Rhee WJ. Detection of exosome miRNAs using molecular beacons for diagnosing prostate cancer. Artif Cells Nanomed Biotechnol 2018;46:S52–S63.10.1080/21691401.2018.1489263Search in Google Scholar PubMed
93 Rahman M, Sampad MJN, Hawkins A, Schmidt H. Recent advances in integrated solid-state nanopore sensors. Lab Chip 2021;21:3030–3052.10.1039/D1LC00294ESearch in Google Scholar
94 Rockett TW, Almahyawi M, Ghimire ML, Jonnalagadda A, Tagliaferro V, Seashols-Williams SJ, Bertino MF, Caputo GA, Reiner JE. Cluster-Enhanced Nanopore Sensing of Ovarian Cancer Marker Peptides in Urine. ACS Sens 2024;9:860–869.10.1021/acssensors.3c02207Search in Google Scholar PubMed PubMed Central
95 Beamish E, Tabard-Cossa V, Godin M. Digital counting of nucleic acid targets using solid-state nanopores. Nanoscale 2020;12:17833–17840.10.1039/D0NR03878DSearch in Google Scholar PubMed
96 Olmedillas-López S, García-Arranz M, García-Olmo D. Current and emerging applications of droplet digital pcr in oncology. Mol Diagn Ther 2017;21:493–510.10.1007/s40291-017-0278-8Search in Google Scholar PubMed
97 Cho SM, Shin S, Kim Y, Song W, Hong SG, Jeong SH, et al. A novel approach for tuberculosis diagnosis using exosomal DNA and droplet digital PCR. Clin Microbiol Infect 2020;26:942.e1–942.e5.10.1016/j.cmi.2019.11.012Search in Google Scholar PubMed
98 Kim Y, Shin S, Lee KA. Exosome-based detection of EGFR T790M in plasma and pleural fluid of prospectively enrolled non-small cell lung cancer patients after first-line tyrosine kinase inhibitor therapy. Cancer Cell Int 2021;21:50.10.1186/s12935-021-01761-xSearch in Google Scholar PubMed PubMed Central
99 Zhang T, Tian T, Zhou R, Li S, Ma W, Zhang Y, et al. Design, fabrication and applications of tetrahedral DNA nanostructure-based multifunctional complexes in drug delivery and biomedical treatment. Nat Protoc 2020;15:2728–2757.10.1038/s41596-020-0355-zSearch in Google Scholar PubMed
100 Gao J, Zhang H, Wang Z. A DNA tetrahedron nanoprobe-based fluorescence resonance energy transfer sensing platform for intracellular tumor-related miRNA detection. Analyst 2020;145:3535–3542.10.1039/C9AN02610JSearch in Google Scholar
101 Fish, K. N. Total internal reflection fluorescence (TIRF) microscopy. Curr Protoc Cytom 2009;Chapter 12:Unit12.18.10.1002/0471142956.cy1218s50Search in Google Scholar PubMed PubMed Central
102 He D, Wang H, Ho SL, Chan HN, Hai L, He X, et al. Total internal reflection-based single-vesicle in situ quantitative and stoichiometric analysis of tumor-derived exosomal microRNAs for diagnosis and treatment monitoring. Theranostics 2019;9:4494–4507.10.7150/thno.33683Search in Google Scholar PubMed PubMed Central
103 Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, et al. Nanoplasmonic Quantification of tumor-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat Biomed Eng 2017;1:0021.10.1038/s41551-016-0021Search in Google Scholar PubMed PubMed Central
104 Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun 2015;6:6999.10.1038/ncomms7999Search in Google Scholar PubMed PubMed Central
105 Zhang P, Wu X, Gardashova G, Yang Y, Zhang Y, Xu L, et al. Molecular and functional extracellular vesicle analysis using nanopatterned microchips monitors tumor progression and metastasis. Sci Transl Med 2020;12:eaaz2878.10.1126/scitranslmed.aaz2878Search in Google Scholar PubMed PubMed Central
106 Hu J, Sheng Y, Kwak KJ, Shi J, Yu B, Lee LJ. A signal-amplifiable biochip quantifies extracellular vesicle-associated RNAs for early cancer detection. Nat Commun 2017;8:1683.10.1038/s41467-017-01942-1Search in Google Scholar PubMed PubMed Central
© 2025 Claudia Wing Lam Tam, Judy Wai Ping Yam, published by De Gruyter on behalf of the SMP
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Articles in the same Issue
- Perspective
- Advances in the diagnosis and treatment of gastrointestinal tumors under the concept of super minimally invasive surgery
- Review Article
- Harnessing the potential of small extracellular vesicle biomarkers for cancer diagnosis and prognosis with advanced analytical technologies
- Current understanding and controversy on brain access of GLP-1 and GLP-1 receptor agonists
- Mitochondrial quality control as a therapeutic target in cardiovascular disease: Mechanistic insights and future directions
- Original Article
- Causal association between gut microbiota composition and the risk of atrial fibrillation
- Artificial intelligence-based predictive model for relapse in acute myeloid leukemia patients following haploidentical hematopoietic cell transplantation
- CircPLK1 upregulates ETS1 to confer anthracycline resistance in triple-negative breast cancer
- N-acetylglucosaminyltransferase V attenuates myocardial infarction by mediating the insulin-like growth factor 1 receptor signaling pathway
- A similar effect of fibrinogen on efficacy and safety of tenecteplase versus alteplase in acute ischemic cerebrovascular events (TRACE II) trial
Articles in the same Issue
- Perspective
- Advances in the diagnosis and treatment of gastrointestinal tumors under the concept of super minimally invasive surgery
- Review Article
- Harnessing the potential of small extracellular vesicle biomarkers for cancer diagnosis and prognosis with advanced analytical technologies
- Current understanding and controversy on brain access of GLP-1 and GLP-1 receptor agonists
- Mitochondrial quality control as a therapeutic target in cardiovascular disease: Mechanistic insights and future directions
- Original Article
- Causal association between gut microbiota composition and the risk of atrial fibrillation
- Artificial intelligence-based predictive model for relapse in acute myeloid leukemia patients following haploidentical hematopoietic cell transplantation
- CircPLK1 upregulates ETS1 to confer anthracycline resistance in triple-negative breast cancer
- N-acetylglucosaminyltransferase V attenuates myocardial infarction by mediating the insulin-like growth factor 1 receptor signaling pathway
- A similar effect of fibrinogen on efficacy and safety of tenecteplase versus alteplase in acute ischemic cerebrovascular events (TRACE II) trial

