Clinical utility of chromosomal microarray and whole exome sequencing in evaluating genetic causes for pregnancy loss using products of conception specimens
-
Kanaka Durga Devi Yadam Reddy
Abstract
Objectives
To determine the genetic causes of miscarriage by analyzing products of conception (POC).
Methods
Chromosomal microarray (CMA) using the Affymetrix Cytoscan HD array was performed in 172 POC specimens from women experiencing spontaneous miscarriage before 20 weeks of gestation to detect aneuploidies, copy number variants (CNVs), and loss of heterozygosity (LOH). Whole exome sequencing (WES) with Roche KAPA HyperExome V2 probes was used for cases where CMA results were normal.
Results
Common clinical indications included recurrent pregnancy loss, first-time miscarriage, absence of cardiac activity, intrauterine death, and fetal growth restriction (FGR), making up 72.55 % of cases. CMA identified chromosomal abnormalities in 38.37 % of samples, with numerical anomalies in 16.86 % and structural anomalies in 21.51 %. Turner syndrome (5.8 %) and various trisomies (5.8 %) were frequent numerical anomalies. Mosaicism and LOH were observed in 11.04 and 2.91 % of cases. WES detected pathogenic or likely pathogenic mutations in 21 genes (e.g., KCNQ1, KCNE1, COL1A2, ROBO1) in 18 cases, adding a 10.46 % diagnostic yield. K-means clustering grouped 17 of these genes into three pathways: chondrocyte differentiation, fibrin clot formation, and Ehlers-Danlos syndrome.
Conclusions
Combining CMA and WES provides a diagnostic yield of 48.83 %, offering a powerful approach to uncover genetic causes of pregnancy loss and guide clinical care.
Introduction
Loss of pregnancy before viability, known as miscarriage, affects approximately 23 million individuals annually, imposing significant physical, physiological, and economic burdens [1]. Chromosomal abnormalities, particularly numerical abnormalities followed by structural aberrations, are the leading causes of miscarriage [2]. The incidence rises sharply with maternal age, with women aged 45 yrs and above experiencing 53 % miscarriage rate compared to 10 % for those aged 25–29 yrs [3]. Paternal age over 45 yrs also increases the risk by 1.75-fold [4]. Genetic analysis techniques, including karyotyping and chromosomal microarray (CMA), show 86 % concordance in identifying chromosomal anomalies. However, CMA offers a 13 % higher diagnostic yield and Karyotyping detects an additional 3 % of unique defects, highlighting their complementary nature [5]. CMA analysis of products of conception (POC) from 65,333 miscarriages identified 57.5 % aneuploidies [6], while exome sequencing proved informative in 22 % of cases [7]. CMA demonstrates superior diagnostic yield in early pregnancy loss compared to late pregnancy loss [8]. The rate of chromosomal abnormalities causing miscarriage in assisted reproductive technology was shown to be around 67 % [9].
Karyotyping fails in 20–40 % of POC samples, whereas SNP-based CMA succeeds in over 90 % [10]. A recent study showed that karyotyping and Fluorescent in situ hybridization (FISH) detect 33.33 % of abnormalities, while array comparative genomic hybridization (aCGH) identifies 24.67 %-primarily copy number variations (CNVs) [11]. Maternal cell contamination, a common issue in chorionic villi analysis, affects 15 % of samples [12].
This study aims to elucidate the genetic underpinnings of miscarriage by analyzing 172 products of conception (POC) specimens using chromosomal microarray (CMA) and whole exome sequencing (WES) with the Roche KAPA HyperExome V2 Probes. The objectives include identifying chromosomal abnormalities, such as aneuploidies, copy number variants, and loss of heterozygosity, through CMA, and characterizing rare, pathogenic mutations in genes associated with embryonic development in cases where CMA is non-informative via WES. Specifically, we seek to determine the contribution of mutations to miscarriage, enhance the diagnostic yield for miscarriage etiology, and establish a genetic profile to inform clinical management and counseling for recurrent pregnancy loss.
Materials and methods
Study design
This retrospective study examined POC samples from women experiencing spontaneous miscarriage between August 2024 and March 2025. Inclusion criteria were POC samples from singleton pregnancies with miscarriage before 20 weeks of gestation, excluding samples with maternal cell contamination or poor DNA quality. Samples were collected from all eligible patients representing pregnancy loss during the study period referred by different gynecologists from Hyderabad for genetic analysis. Of 193 POC samples collected with informed consent, 172 were analyzed after exclusions. The study was approved by the Institutional Ethics Committee of Erode Cancer Centre (IEC-ECC/2024/MGRBT/PhD/23–24/001-001).
Sample collection and processing
POC specimens were obtained via dilation and curettage (D&C) or natural expulsion, placed in RPMI-1640 with 10 % fetal bovine serum, and transported to the laboratory within 2 h at 4 °C. Chorionic villi were dissected under a stereomicroscope to minimize maternal tissue contamination. DNA was extracted using QIAamp DNA Mini Kit, Qiagen according to the manufacturer’s protocol. DNA quantity and quality were assessed using Nanodrop spectrophotometer and Qubit fluorometer.
Chromosomal microarray analysis (CMA)
CMA was performed using an Affymetric CytoScan HD array (50–100 kB resolution). About 250 ng of DNA was fragmented, cy3 labeled, and hybridized to microarray according to the manufacturer’s instructions. Arrays were scanned with a GeneChip Scanner 3,000 and data were analyzed with Chromosome Analysis Suite (ChAS) to identify CNVs, aneuploidies, and regions of homozygosity. Results were interpreted based on the Database of Genomic Variants and confirmed by QF-PCR or FISH for ambiguous findings.
Whole exome sequencing (WES)
Library preparation
WES libraries were prepared using the Roche KAPA HyperExome V2 Probes via the KAPA HyperCap Evolved Workflow v4. A 100 ng of genomic DNA was enzymatically sheared to 150–200 bp using KAPA EvoPrep or KAPA EvoPlus V2 Kit, end-repaired, A-tailed, and ligated to indexed adapters. The adapter-ligated DNA was amplified via pre-capture PCR to enrich exonic regions. Targeted capture was performed using the Roche KAPA HyperExome V2 Probes, which target approximately 43 Mb of exonic regions, followed by hybridization and streptavidin bead-based enrichment. Post-capture PCR amplified the library.
Sequencing
Enriched libraries were quantified using qPCR and sequenced on an MGI DNBSEQ-G50 platform, generating 150 bp paired-end reads. The target depth was 100–150x, with at least 90 % of targeted bases covered at 20x.
Data processing and variant calling
FASTQ files were quality-checked with FastQC. Reads were aligned to the human reference genome (GRCh38) using the Burrows-Wheeler Aligner (BWA-MEM), duplicate reads were removed with Picard Tools, and base quality recalibrated with Genome Analysis Toolkit (GATK). Variants were called using GATK HaplotypeCaller, joint-genotyped, and filtered with Variant quality score recalibration (VQSR).
Variant annotation and filtering
Variants were annotated with ANNOVAR or Ensembl Variant Effect Predictor (VEP) to identify functional consequences (e.g., missense, nonsense, splice-site variants) and population frequencies from databases such as gnomAD, ExAC, and 1,000 Genomes. Variants with a minor allele frequency (MAF) >1 % were excluded unless clinically relevant. Pathogenicity was assessed using SIFT, PolyPhen-2, CADD and databases like ClinVar and HGMD, prioritizing based on inheritance, gene function, and clinical relevance.
Validation
High-priority variants were confirmed by Sanger sequencing or targeted amplicon sequencing, including segregation analysis where applicable.
Quality control
Samples were excluded if maternal cell contamination exceeded 15 %, as determined by STR profiling, or if DNA integrity was compromised (e.g., OD 260/280 ratio <1.8). Positive and negative controls were included in each CMA and WES and sequencing run to validate results.
Quality control metrics were monitored at each step, including DNA integrity, library fragment size distribution, capture efficiency, and sequencing coverage. Samples with <80 % of targeted bases at 20x coverage were re-sequenced.
Gene clustering analysis
Candidate genes were selected from the 21 mutations identified by WES in 18 POC samples (Table 1) and annotated using Gene Ontology terms for biological pathways. K-means clustering (k=3) was performed using Euclidean distance in R software, with cluster validity assessed by silhouette score (average score: 0.62, indicating good separation). The analysis grouped genes into three clusters based on shared functional pathways relevant to pregnancy loss. A network diagram was generated using Cytoscape to visualize clusters and their relationships.
Key mutations identified through whole exome sequencing in products of conception.
| Case | Genes | Mutation, s | Disease | Clinical history |
|---|---|---|---|---|
| 1 | KCNQ1, KCNE1 | c.1006G>T, p.A336S; c.200G>A, p.R67H | Long QT syndrome | Three missed abortions, one stillbirth |
| 2 | NOTCH3 | c.6097C>T, p.P2033S | Lateral meningocele syndrome | Cleft lip |
| 3 | ARSE | c.17 A>G, p.E6G | X-linked adrenoleukodystrophy (X-ALD) | Polyhydramnios, binder facies, chondrodysplasia punctata |
| 4 | PTPN11 | c.91G>T, p.A31S | Noonan syndrome | IVF pregnancy aborted at 12 weeks 4 days |
| 5 | COL1A2 | c.2594G>T, p.G865V | Osteogenesis imperfecta type 2/3 | USG suggestive of skeletal dysplasia |
| 6 | CTNS | c.422C>T, p.S141F; c.1085+1G>A | Nephropathic cystinosis | Echogenic bowel, moderate oligohydramnios |
| 7 | SERPINC1 | c.1255G>A, p.A419T | Hereditary antithrombin deficiency | Thick NT, cystic hygroma, truncal edema |
| 8 | ACOX2, SLFN14 | c.461_464del, p.T154Sfsa; c.920del, p.G307Vfsa5 | Congenital bile acid synthesis defect; platelet-type bleeding disorder 20 | Recurrent pregnancy loss |
| 9 | COL5A1 | c.4411G>A, p.G1471S | Ehlers-danlos syndrome | Recurrent pregnancy loss |
| 10 | GFI1B | c.551G>C, p.R184P | Platelet-type bleeding disorder 17 | Two miscarriages, absence of cardiac activity |
| 11 | CTNNB1 | c.1154 T>G, p.L385R | CTNNB1-related neurodevelopmental disorder | Left ventricle abnormality, corpus callosum hypoplasia |
| 12 | SOX9 | c.686-1G>A (splicing) | Campomelic dysplasia | Cystic hygroma, skeletal anomalies |
| 13 | F8, KIF20A | c.1018G>A, p.E340K; c.1136G>A, p.R379H | Hemophilia A; familial restrictive cardiomyopathy | Recurrent pregnancy loss, absence of cardiac activity |
| 14 | G6PD | c.949G>A, p.E317K | G6PD deficiency | Intrauterine fetal death, non-immune hydrops |
| 15 | CHD7 | c.3317 A>C, p.E1106A | CHARGE syndrome | Midline cleft lip/palate, cardiac defects |
| 16 | ABCB4, CHD7 | c.808G>C, p.G270R; c.3202-3 T>C (splice region) | Familial intrahepatic cholestasis; CHARGE syndrome | Two miscarriages, absence of cardiac activity |
| 17 | KIF1BP | c.1151del, p.L384a | Goldberg-shprintzen syndrome | Corpus callosum agenesis, cerebellar hypoplasia |
| 18 | ROBO1 | c.1537C>T, p.R513a | Congenital heart disease | Complex cardiac anomaly, single outflow tract |
-
Mutations are heterozygous unless specified (e.g., homozygous for ARSE, G6PD, KIF1BP). Variant classifications (Pathogenic, Likely Pathogenic, VUS) are detailed in the Results section. USG, Ultrasound; NT, nuchal translucency; TIFFA, targeted imaging for fetal anomalies. aCompound heterozygous.
Data analysis
Chromosomal abnormalities were classified as aneuploidies (trisomies, monosomies, polyploidies), copy number variations (e.g., deletions, duplications, gains, losses), mosaic (e.g., mixed cell lines), and normal. Fisher exact test was carried out to establish association of numerical and structural anomalies with specific clinical indications by computing the data in 2 × 2 contingency table format for the presence or absence of anomalies between two groups. The data is represented in the form of odds ratio (OR) and 95 % confidence interval (CI). The p-value of <0.05 was considered statistically significant.
Results
Of 193 POC samples, 172 were analyzed after excluding 11 for maternal cell contamination, 6 for bacterial contamination, and four for poor DNA quality. CMA detected chromosomal abnormalities in 66 samples (38.37 %).
Recurrent pregnancy loss, first-time miscarriage, absence of cardiac activity/intrauterine death (combined as these represent similar diagnoses of fetal demise without distinct clinical differentiation in this study), and fetal growth restriction (FGR) accounted for 72.55 % of cases (Table 2). CMA yields were 45.45 % for recurrent pregnancy loss, 32.14 % for first-time miscarriage, 37.5 % for absence of cardiac activity/intrauterine death, and 33.33 % for FGR.
Clinical indications vs. chromosomal anomalies in the products of conception.
| Indications | Aneuploidies | Copy number variations | Normal CMA | Total cases |
|---|---|---|---|---|
| Recurrent pregnancy loss | 10 | 5 | 11 | 26 |
| First-time miscarriage | 6 | 3 | 19 | 28 |
| Absence of cardiac activity/intrauterine death | 8 | 7 | 25 | 40 |
| Fetal growth restriction | 2 | 4 | 8 | 14 |
| Other | 3 | 8 | 53 | 64 |
| Total | 29 | 27 | 116 | 172 |
-
Aneuploidies were most frequent in recurrent pregnancy loss (RPL, n=10) and miscarriage (n=6), while copy number variations predominated in absence of cardiac activity/intrauterine death (n=7). Aneuploidies were higher in spontaneous miscarriage than other clinical indications (30.19 vs. 13.43 %). Hence, the risk of miscarriage in the presence of aneuploidies was 2.8-fold (95 % CI: 1.12–6.96, p=0.04). Copy number variations (30.12 vs. 17.78 %) were associated with a 5.6-fold higher risk of congenital anomalies (e.g., cardiac, skeletal) compared to aneuploidies (95 % CI: 1.78–17.38, p=0.005).
Aneuploidies were higher in spontaneous miscarriage than other clinical indications (30.19 vs. 13.43 %). Hence, the risk of miscarriage in the presence of aneuploidies was 2.8-fold (95 % CI: 1.12–6.96, p=0.04). Copy number variations (30.12 vs. 17.78 %) were associated with a 5.6-fold higher risk of congenital anomalies (e.g., cardiac, skeletal) compared to aneuploidies (95 % CI: 1.78–17.38, p=0.005). CMA was not informative for two cases with Chiari-2 malformation and cleft lip/palate.
Turner syndrome was observed in 10 POC samples (5.8 %) and Trisomies were reported in 10 samples (5.8 %), including Trisomy 21 (n=3), Trisomy 16 (n=3), and Trisomy 13 (n=1), Trisomy 7 (n=1), Trisomy 18 (n=1), and Trisomy 22 (n=1). Polyploidy and triploidy (Figure 1) each occurred in 3 samples (1.7 %). Loss of heterozygosity (LOH) was seen in five cases (2.9 %).

Chromosome analysis suite (ChAS) analysis showing triploidy in products of conception. Chromosomal microarray (CMA) output from the Affymetrix CytoScan HD array analyzed using the Chromosome Analysis Suite (ChAS) software, showing triploidy in a product of conception (POC) sample. The X-axis is representative of chromosomes. The plot displays four allele peaks across all chromosomes (indicated by distinct banding patterns), consistent with a triploid karyotype (69,XXX or 69,XXY). This finding reflects three copies of each chromosome, a common genetic cause of early miscarriage. Note: The X-axis represents chromosomes 1–22 (autosomes); sex chromosomes may show similar patterns but are not labeled here.
Deletions were observed in 5 samples (2.9 %): 15q11.2 del, 2q24.2 del, del Chr1/dup chr3, del Chr2 20 MB, Del Chr8/Gain mosaic X. Duplications were reported in 2specimens (1.16 %): DupChr3/del Chr1, dup X43.82 Mb.
Gains affected seven samples (4.1 %): 22q12.3 Gain 0.64 Mb, 22q13 Gain, Chr17 gain 0.67 Mb, Gain 11, Loss 16, X, Gain 22. Losses affected 6 samples (3.5 %): loss16, X, loss mosaicism 7, 19, loss chr16 1.16 Mb, loss X, loss16 chr 0.67 mb. One sample showed multiple gains and losses.
Whole exome sequencing in 18 cases where CMA is not informative revealed the contribution of mutations in 21 genes in miscarriage (KCNQ1, KCNE1, NOTCH3, ABCD1, PTPN11, COL1A2, CTNS, SERPINC1, ACOX2, SLFN14, COL5A1, GF1B, CTNNB1, SOX9, F8, G6PD, CHD7, ARSE, KIF1B, ROBO1). Whole exome sequencing increases the diagnostic yield by 10.46 %. (Table 1). Among these, all the mutations are pathogenic or likely pathogenic except for NOTCH3, ARSE, CTNS, KIF20A and CHD7 variants that are variants of uncertain significance.
Three samples required re-sequencing due to initial coverage below 80 % of targeted bases at 20x, representing a re-sequencing rate of 1.74 % (3/172).
K-means clustering of 17 candidate genes from WES data revealed three distinct clusters (Figure 2). Cluster 1, comprising CHD7, SOX9, NOTCH3, PTPN11, CTNNB1, GFI1B, ROBO1, KIF20A, KCNQ1, and KCNE1 (10 genes), was associated with negative regulation of chondrocyte differentiation, potentially linked to skeletal anomalies observed in POC samples (e.g., SOX9 with cystic hygroma). Cluster 2, including SERPINC1, F8, G6PD, ACOX2, and ABCB4 (5 genes), was tied to fibrin clot formation, suggesting placental vascular contributions (e.g., SERPINC1 with thick nuchal translucency). Cluster 3, with COL5A1 and COL1A2 (2 genes), aligned with Ehlers-Danlos syndrome, consistent with skeletal dysplasia findings (e.g., COL1A2 case).
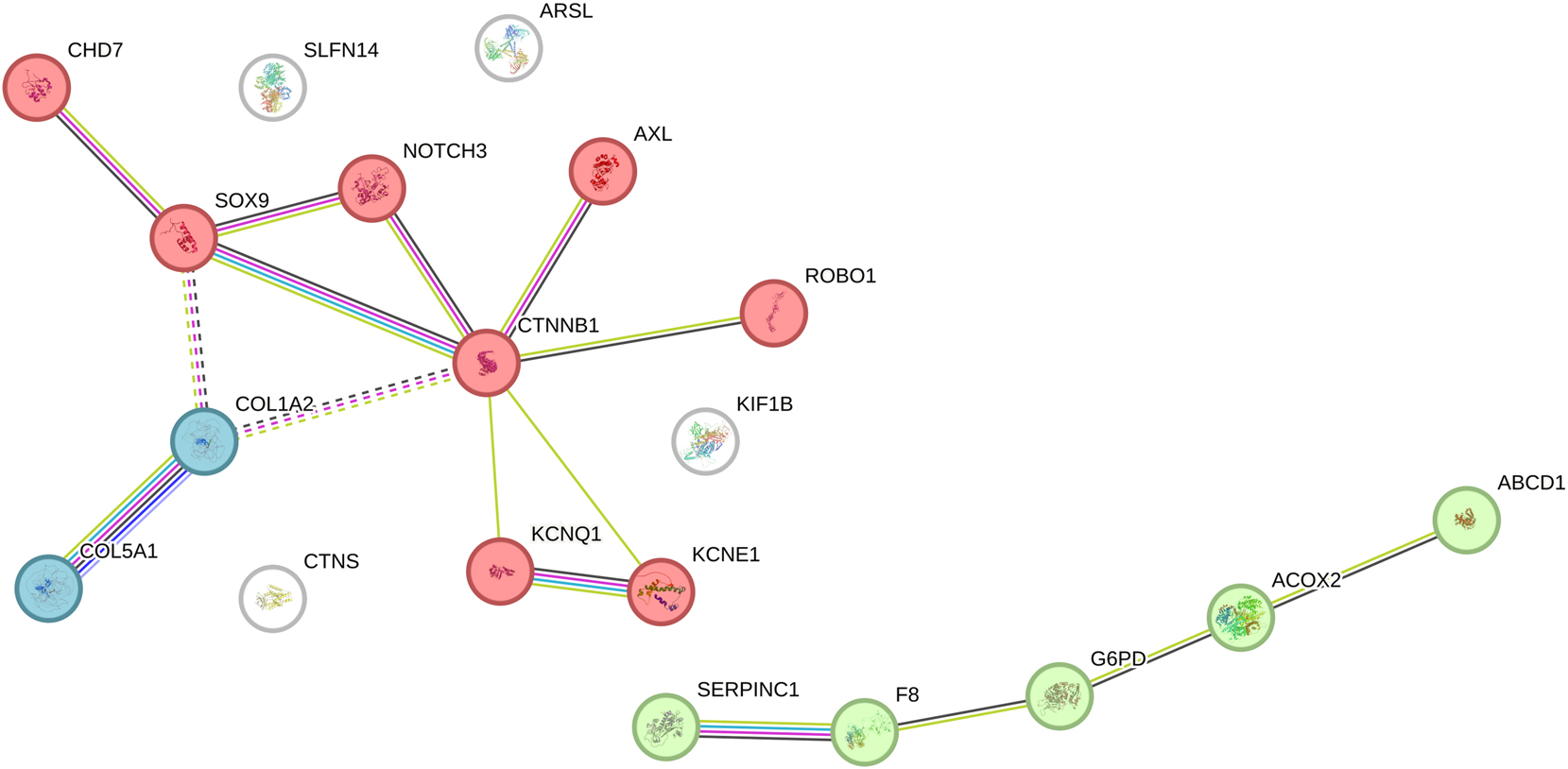
K-mean clustering of candidate genes associated with pregnancy loss. Network diagram of K-means clustering (k=3) applied to 17 candidate genes identified from whole exome sequencing (WES) data, based on functional annotations. Cluster 1 (red nodes: CHD7, SOX9, NOTCH3, PTPN11, CTNNB1, GFI1B, ROBO1, KIF20A, KCNQ1, KCNE1) includes 10 genes associated with negative regulation of chondrocyte differentiation, potentially linked to skeletal and developmental defects. Cluster 2 (green nodes: SERPINC1, F8, G6PD, ACOX2, ABCB4) includes 5 genes tied to the common pathway of fibrin clot formation, suggesting roles in placental vascular issues. Cluster 3 (blue nodes: COL5A1, COL1A2) includes 2 genes associated with Ehlers-Danlos syndrome, reflecting connective tissue abnormalities. Edges indicate functional relationships based on pathway annotations. White nodes are not part of clustering.
Discussion
In the current study, we demonstrated 38.37 % chromosomal abnormalities in 172 products of conception by CMA. Recurrent pregnancy loss, first-time miscarriage, absence of cardiac activity/intrauterine death, and intrauterine growth restrictions are the major clinical indications in this cohort. Aneuploidies and copy number variations were observed in 16.86 and 21.51 % of products of conception by CMA. Among the aneuploidies by CMA, Turner syndrome and different trisomies are the major contributors to pregnancy loss followed by triploidy and polyploidy. Mosaicism was observed in 11.04 % of cases by CMA. Gain or loss of chromosomes accounted for 7.56 % of cases by CMA. Deletions and duplications accounted for 4.07 % of cases, as determined by CMA. Pathogenic or likely pathogenic genetic variants were found to contribute to 10.46 % of pregnancy losses by WES.
The incidence of chromosomal abnormalities was reported to be lower in subjects with recurrent pregnancy loss than those with sporadic pregnancy loss [13]. However, we observed 45.45 % of chromosomal abnormalities in the recurrent pregnancy loss cases in this study by CMA. ArrayCGH can detect 35 % of chromosomal abnormalities even in products of conception culture failure specimens [14].
Our study is in agreement with Matuszewska et al. in demonstrating Turner syndrome and trisomies as the most common chromosomal abnormalities in the spontaneous miscarriage samples [2]. A recent study of 1160 POC samples reported 45.8 % single aneuploidies, 2.7 % multiple aneuploidies, 4.3 % polyploidies, 4.7 % partial aneuploidies, and 6.6 % submicroscopic CNVs in early miscarriages [15]. In another study of 840 chorionic villi samples from spontaneous abortion, 37.35 % were aneuploidies and 6.97 % were polyploidies [16].
Although triploidy is one of the most common causes of early miscarriage, rarely few fetuses survive into second-trimester pregnancy and exhibit intrauterine growth restriction, oligohydramnios, bilateral cerebral ventriculomegaly, structural heart defects, and Dandy-Walker malformations [17]. Carriers of structural chromosomal defects were reported to exhibit recurrent spontaneous miscarriage, oligospermia, azoospermia, primary amenorrhea, and fetal death [18].
The current study reported three spontaneous abortions in a 31-yr-old female with one of the POC samples exhibiting KCNQ1 and KCNE1 heterozygous mutations associated with Long QT syndrome. Kasak et also reported KCNQ1 mutation in the mother and two POC specimens suggesting the association of this mutation with pregnancy loss [19]. This is further confirmed by another study of 64 pregnancies from 23 women with long QT syndrome exhibiting 40.6 % pregnancy losses [20]. COL1A1/A2 mutations were reported to be more common in fetuses with skeletal abnormalities [21].
A study of 22 pregnancies in 8 women homozygous for SERPINC1 revealed adverse pregnancy outcomes in 68.18 % of pregnancies including an equal number of early pregnancy losses and intrauterine deaths [22]. COL1A1, COL1A2, and COL5A2 are associated with preterm premature rupture of membranes and cervical incompetence [23]. The differential expression of CDK11A, C19orf71, COL5A1, and GNE in advanced maternal age was reported to increase the risk for spontaneous abortion [24].
A woman carrying balanced reciprocal translocation 46, XX, t (7; 17) (p13; q24) involving SOX9 exhibited IVF pregnancies affected with aneuploidies and also had miscarriages even with donor oocytes [25]. G6PD deficiency was reported to increase the risk for nonimmune hydrops fetalis (NIHF) and several fetal anemia, which in turn increases the risk of miscarriage [26].
KIF1BP is a regulator of a kinesin subset essential for neural development and most of the mutations observed in Goldberg-Shprintzen syndrome are nonsense mutations associated with loss of function [27]. They were reported to contribute to defects in neuronal migration, morphogenesis, maturation, and survival [27].
Li et al. reported reduced β-hCG and trophoblast invasion in human placenta samples from women with missed and threatened miscarriages with abnormal expression of Slit2 and Robo1 [28]. They postulated that blocking SLIT2/ROBO1 signaling affects trophoblast differentiation and angiogenesis-related gene expression thus contributing to increased risk for miscarriage [28]. We reported ROBO1 mutation in a POC sample causing spontaneous abortion.
The K-means clustering (Figure 2) provided functional insights into the 17 WES-identified genes, grouping them into three pathways. Cluster 1 (e.g., SOX9, CHD7) suggests chondrocyte differentiation defects may contribute to skeletal anomalies in miscarriage, supported by cases like SOX9-related skeletal defects and COL1A2-associated dysplasia. Cluster 2 (e.g., SERPINC1, F8) indicates fibrin clot formation issues, potentially affecting placental integrity, aligning with SERPINC1’s adverse pregnancy outcomes [22]. Cluster 3 (COL5A1, COL1A2) reinforces connective tissue roles, consistent with Ehlers-Danlos syndrome and prior reports [23]. The inclusion of KCNQ1 and KCNE1 in Cluster 1 may reflect indirect skeletal effects via cardiac dysfunction, as Long QT syndrome can impact fetal development [19], warranting further investigation. This pathway analysis enhances the 48.83 % diagnostic yield, highlighting diverse genetic mechanisms for future study.
This study’s retrospective design may introduce selection bias, and the lack of parental genetic data limits the ability to distinguish de novo from inherited variants. WES focused on exonic regions, potentially missing intronic or regulatory variants. Future studies should incorporate trio sequencing and larger RPL cohorts.
To conclude, this study demonstrates the complementary roles of CMA and WES in elucidating genetic causes of miscarriage. CMA identified chromosomal abnormalities in 38.37 % of 172 POC samples, with Turner syndrome and trisomies predominant, while WES revealed pathogenic mutations in 21 genes (e.g., KCNQ1, ROBO1) in 22.3 % of non-informative cases (Figure 3). These findings highlight diverse genetic mechanisms underlying pregnancy loss and underscore the importance of integrated genetic testing to improve diagnostic yield and guide clinical management for recurrent pregnancy loss.

Graphical Abstract: CMA identified chromosomal abnormalities in (66/172) 38.37 % of samples, with aneuploidies in 16.86 % (29/172) and copy number variations in 21.51 % (37/172). The copy number variations were: mosaicism (n=16), LOH (n=4), losses (n=5), gains (n=6), duplications (n=2), deletions (n=4). The classical aneuploidies were: Turner syndrome (n=10), trisomies (n=10), triploidy (n=3), polyploidy (n=3). Out of the 172 POC samples tested for WES, only 18 samples had pathogenic or likely pathogenic mutations in 21 genes (e.g., KCNQ1, KCNE1, COL1A2, ROBO1), adding a 10.46 % diagnostic yield. K-means clustering grouped 17 of these genes into three pathways: chondrocyte differentiation, fibrin clot formation, and Ehlers-Danlos syndrome.
Acknowledgment
We thank all the families who participated in the study.
-
Research ethics: The study protocol was approved by the Institutional Ethics Committee of Erode Cancer Centre (IEC-ECC/2024/MGRBT/PhD/23–24/001-001).
-
Informed consent: Informed consent was obtained from all individuals included in this study, or their legal guardians or wards.
-
Author contributions: SMN, KS and TH conceptualized and designed the study. KDYR, performed the wet lab experiments related to whole exome sequencing and chromosomal microarray. PP performed bioinformatic analysis. SMN and TH genotyped and interpreted the results. Manuscript was drafted by SMN and critically revised by KS and TH. All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Use of Large Language Models, AI and Machine Learning Tools: The English language editing was done using Grok AI tool.
-
Conflict of interest: The authors state no conflict of interest.
-
Research funding: The study was conducted using intramural grants provided by the management of Yoda diagnostics Pvt Lt.
-
Data availability: Not applicable.
References
1. Quenby, S, Gallos, ID, Dhillon-Smith, RK, Podesek, M, Stephenson, MD, Fisher, J, et al.. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet 2021;397:1658–67. https://doi.org/10.1016/S0140-6736(21)00682-6.Suche in Google Scholar PubMed
2. Matuszewska, KE, Bukowska-Olech, E, Piechota, M, Staniek-Łacna, K, Drews, K, Więckowska, B, et al.. From chromosomal aberrations to mutations in individual genes – the significance of genetic studies of chorions after miscarriage in the search for causes of miscarriages. J Matern Fetal Neonatal Med 2024;37:2364249. https://doi.org/10.1080/14767058.2024.2364249.Suche in Google Scholar PubMed
3. Magnus, MC, Wilcox, AJ, Morken, NH, Weinberg, CR, Håberg, SE. Role of maternal age and pregnancy history in risk of miscarriage: prospective register based study. BMJ 2019;364:l869. https://doi.org/10.1136/bmj.l869.Suche in Google Scholar PubMed PubMed Central
4. du Fossé, NA, van der Hoorn, MP, van Lith, JMM, le Cessie, S, Lashley, EELO. Advanced paternal age is associated with an increased risk of spontaneous miscarriage: a systematic review and meta-analysis. Hum Reprod Update 2020;26:650–69. https://doi.org/10.1093/humupd/dmaa010.Suche in Google Scholar PubMed PubMed Central
5. Dhillon, RK, Hillman, SC, Morris, RK, McMullan, D, Williams, D, Coomarasamy, A, et al.. Additional information from chromosomal microarray analysis (CMA) over conventional karyotyping when diagnosing chromosomal abnormalities in miscarriage: a systematic review and meta-analysis. BJOG 2014;121:11–21. https://doi.org/10.1111/1471-0528.12382.Suche in Google Scholar PubMed
6. Kutteh, WH, Papas, RS, Maisenbacher, MK, Dahdouh, EM. Role of genetic analysis of products of conception and PGT in managing early pregnancy loss. Reprod Biomed Online 2024;49:103738. https://doi.org/10.1016/j.rbmo.2023.103738.Suche in Google Scholar PubMed
7. Zhao, C, Chai, H, Zhou, Q, Wen, J, Reddy, UM, Kastury, R, et al.. Exome sequencing analysis on products of conception: a cohort study to evaluate clinical utility and genetic etiology for pregnancy loss. Genet Med 2021;23:435–42. https://doi.org/10.1038/s41436-020-01008-6.Suche in Google Scholar PubMed
8. Gou, L, Liu, T, Wang, Y, Wu, Q, Hu, S, Dong, B, et al.. Clinical utilization of chromosomal microarray analysis for the genetic analysis in subgroups of pregnancy loss. J Matern Fetal Neonatal Med 2022;35:4404–11. https://doi.org/10.1080/14767058.2020.1849126.Suche in Google Scholar PubMed
9. Kato, T, Miyai, S, Suzuki, H, Murase, Y, Ota, S, Yamauchi, H, et al.. Usefulness of combined NGS and QF-PCR analysis for product of conception karyotyping. Reprod Med Biol 2022;21:e12449. https://doi.org/10.1002/rmb2.12449.Suche in Google Scholar PubMed PubMed Central
10. Sahoo, T, Dzidic, N, Strecker, MN, Commander, S, Travis, MK, Doherty, C, et al.. Comprehensive genetic analysis of pregnancy loss by chromosomal microarrays: outcomes, benefits, and challenges. Genet Med 2017;19:83–9. https://doi.org/10.1038/gim.2016.69.Suche in Google Scholar PubMed
11. Gajjar, K, Patel, A, Patel, B, Chettiar, S, Jhala, D. Array comparative genomic hybridization analysis of products of conception in recurrent pregnancy loss for specific anomalies detected by USG. Reprod Fertil 2023;4:e220092. https://doi.org/10.1530/RAF-22-0092.Suche in Google Scholar PubMed PubMed Central
12. Ouchi, N, Takeshita, T, Kasano, S, Yokote, R, Yonezawa, M, Kurashina, R, et al.. Maternal cell contamination in embryonic chromosome analysis of missed abortions. J Obstet Gynaecol Res 2022;48:1641–7. https://doi.org/10.1111/jog.15249.Suche in Google Scholar PubMed
13. Lei, D, Zhang, XY, Zheng, PS. Recurrent pregnancy loss: fewer chromosomal abnormalities in products of conception? A meta-analysis. J Assist Reprod Genet 2022;39:559–72. https://doi.org/10.1007/s10815-022-02414-2.Suche in Google Scholar PubMed PubMed Central
14. Zhou, Q, Wu, SY, Amato, K, DiAdamo, A, Li, P. Spectrum of cytogenomic abnormalities revealed by array comparative genomic hybridization on products of conception culture failure and normal karyotype samples. J Genet Genomics 2016;43:121–31. https://doi.org/10.1016/j.jgg.2016.02.002.Suche in Google Scholar PubMed
15. Xue, H, Guo, Q, Yu, A, Lin, M, Chen, X, Xu, L, et al.. Genetic analysis of chorionic villus tissues in early missed abortions. Sci Rep 2023;13:21719. https://doi.org/10.1038/s41598-023-48358-0.Suche in Google Scholar PubMed PubMed Central
16. Jia, CW, Wang, L, Lan, YL, Song, R, Zhou, LY, Yu, L, et al.. Aneuploidy in early miscarriage and its related factors. Chin Med J (Engl). 2015;128:2772–6. https://doi.org/10.4103/0366-6999.167352.Suche in Google Scholar PubMed PubMed Central
17. Blaicher, W, Ulm, B, Ulm, MR, Hengstschläger, M, Deutinger, J, Bernaschek, G, et al.. Dandy-walker malformation as sonographic marker for fetal triploidy. Ultraschall Med 2002;23:129–33. https://doi.org/10.1055/s-2002-25189.Suche in Google Scholar PubMed
18. Wang, HL, Wu, B, Guo, KM, Tian, RH. Psychological characteristics of and counseling for carriers of structural chromosome abnormalities. Genet Mol Res. 2016;15. https://doi.org/10.4238/gmr.15028159.Suche in Google Scholar PubMed
19. Kasak, L, Rull, K, Yang, T, Roden, DM, Laan, M. Recurrent pregnancy loss and concealed long-QT syndrome. J Am Heart Assoc 2021;10:e021236. https://doi.org/10.1161/JAHA.121.021236.Suche in Google Scholar PubMed PubMed Central
20. Albertini, L, Ezekian, J, Care, M, Silversides, C, Sermer, M, Gollob, MH, et al.. Assessment of severity of long QT syndrome phenotype and risk of fetal death. J Am Heart Assoc 2023;12:e029407. https://doi.org/10.1161/JAHA.122.029407.Suche in Google Scholar PubMed PubMed Central
21. Cao, J, Chen, A, Tian, L, Yan, L, Li, H, Zhou, B, et al.. Application of whole exome sequencing in fetal cases with skeletal abnormalities. Heliyon 2022;8:e09819. https://doi.org/10.1016/j.heliyon.2022.e09819.Suche in Google Scholar PubMed PubMed Central
22. Kraft, J, Sunder-Plassmann, R, Mannhalter, C, Quehenberger, P, Tews, G, Langer, M, et al.. Women with homozygous AT deficiency type II heparin-binding site (HBS) are at high risk of pregnancy loss and pregnancy complications. Ann Hematol 2017;96:1023–31. https://doi.org/10.1007/s00277-017-2965-2.Suche in Google Scholar PubMed
23. Anum, EA, Hill, LD, Pandya, A, Strauss, JF3rd. Connective tissue and related disorders and preterm birth: clues to genes contributing to prematurity. Placenta 2009;30:207–15. https://doi.org/10.1016/j.placenta.2008.12.007.Suche in Google Scholar PubMed PubMed Central
24. Qin, M, Chen, W, Hua, L, Meng, Y, Wang, J, Li, H, et al.. DNA methylation abnormalities induced by advanced maternal age in villi prime a high-risk state for spontaneous abortion. Clin Epigenet 2023;15:44. https://doi.org/10.1186/s13148-023-01432-w.Suche in Google Scholar PubMed PubMed Central
25. Peregrino, PFM, Gomes, A, Fujii, M, Bonetti, TCS, Riboldi, M, Monteleone, PAA, et al.. The impact of balanced reciprocal translocation – 46,XX,t(7;17)(p13;q24) probably involving the SOX9 gene in the in vitro fertilization with own oocytes evaluated by preimplantation genetic testing or donated oocytes. JBRA Assist Reprod. 2019;23:68–71. https://doi.org/10.5935/1518-0557.20180066.Suche in Google Scholar PubMed PubMed Central
26. Iyer, NS, Mossayebi, MH, Gao, TJ, Haizler-Cohen, L, Di Mascio, D, McLaren, RAJr, et al.. Glucose-6-phosphate dehydrogenase deficiency as a cause for nonimmune hydrops fetalis and severe fetal anemia: a systematic review. Mol Genet Genomic Med 2024;12:e2491. https://doi.org/10.1002/mgg3.2491.Suche in Google Scholar PubMed PubMed Central
27. Chang, HY, Cheng, HY, Tsao, AN, Liu, C, Tsai, JW. Multiple functions of KBP in neural development underlie brain anomalies in goldberg-shprintzen syndrome. Front Mol Neurosci 2019;12:265. https://doi.org/10.3389/fnmol.2019.00265.Suche in Google Scholar PubMed PubMed Central
28. Li, P, Shi, Y, Shuai, H, Cai, Y, Lu, W, Wang, G, et al.. Altered SLIT2/ROBO1 signalling is linked to impaired placentation of missed and threatened miscarriage in early pregnancy. Histopathology 2017;71:543–52. https://doi.org/10.1111/his.13250.Suche in Google Scholar PubMed
© 2025 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.

