The fetal exposome and Preterm Birth: a systematic synthesis of environmental exposures and multi-omics evidence
-
Wiku Andonotopo
, Muhammad Adrianes Bachnas
, Julian Dewantiningrum
Abstract
Objectives
Preterm birth (PTB), defined as delivery before 37 weeks of gestation, is a leading cause of neonatal mortality and long-term developmental impairment. Its complex etiology, spanning environmental, genetic, psychosocial, and socio-economic domains, limits effective prediction and prevention. We systematically synthesized evidence on how environmental exposures influence PTB risk through multi-omic disruptions within a fetal exposome framework.
Methods
A comprehensive literature search was conducted in major biomedical databases, following PRISMA guidelines. Ninety-five human studies published through May 2025 were included, encompassing exposures such as ambient air pollution, endocrine-disrupting chemicals, maternal stress, nutrition, occupational hazards, climate variability, and microbiome alterations. Two reviewers independently extracted data (exposure type, omics platform, biospecimen, PTB subtype) with inter-rater reliability assessment, and study quality was evaluated using the Newcastle–Ottawa Scale. Findings were narratively stratified by exposure category, study design, and spontaneous vs. indicated PTB.
Results
Environmental exposures were consistently associated with disruptions in oxidative stress, inflammation, immune regulation, hormonal signaling, placental aging, and microbial ecology, mediated by multi-omic signatures in maternal, placental, and fetal tissues. Candidate biomarkers show promise for early risk stratification but lack validation and population-level predictive performance due to heterogeneous exposure assessment and study design.
Conclusions
Integrating fetal exposome concepts with multi-omics enhances mechanistic insight into PTB risk and may support biomarker discovery and precision-guided prenatal interventions. Clinical translation requires standardized exposure measurement, biomarker validation, and equity-focused implementation.
Introduction
Preterm birth (PTB), defined as delivery before 37 weeks of gestation, remains the leading cause of neonatal mortality and long-term developmental impairment worldwide [1], [2], [3], [4]. Its etiology is multifactorial, driven by genetic, environmental, psychosocial, and socio-structural determinants . Conventional risk prediction models, largely based on obstetric history and demographic factors, fail to account for upstream environmental and social determinants that influence pregnancy outcomes [9], [10], [11], [12], [13], [14], [15].
The fetal exposome – the totality of in utero exposures from conception through delivery – provides a systems-level framework to evaluate PTB risk [16], [17], [18], [19], [20], [21], [22]. This framework includes chemical exposures (e.g., air pollutants, endocrine-disrupting chemicals, metals, nanoplastics) and nonchemical stressors (e.g., maternal stress, structural racism, nutrition, occupational hazards, climate variability, and microbiome alterations) [17], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37]. These factors influence biological pathways associated with inflammation, oxidative stress, immune dysregulation, endocrine imbalance, and placental aging, all of which contribute to increased PTB susceptibility [20], 28], 30], [32], [33], [34, [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54]. Key environmental exposures and their biological effects are summarized in Table 1.
Summary of key environmental exposures linked to preterm birth with NOS quality grading.
| Authors | Exposure type | Main sources | Associated omic changes | Insights, strengths, limitations | NOS quality (0–9) |
| Burris et al. 2016, EHP [1] | Air pollution (PM2.5) | Traffic emissions, urban smog | Placental inflammation, oxidative stress, DNA methylation in immune pathways | Strong causal inference; human cohort with precise exposure windows. Limited to urban US setting. | 8 |
| Ding et al. 2023, Environ Int [4] | Air pollution (PM2.5, NO2) | Urban air, Beijing | Altered lipid metabolism, oxidative stress in placenta | Prospective cohort; trimester-specific data. Limited omic depth beyond metabolism. | 7 |
| Vrijheid et al. 2014, EHP [17] | Air pollution (HELIX study) | Urban environments (Europe) | DNA methylation: angiogenesis, immune genes | Landmark multi-country design, rich omic data. Less racial/ethnic diversity. | 8 |
| La Merrill et al. 2023, Environ Res [10] | EDCs | Phthalates, BPA, PFAS | Transcriptomic dysregulation, receptor gene methylation | Broad toxicant scope. Cross-sectional design limits temporal inference. | 6 |
| Fang et al. 2024, Environ Pollut [21] | EDCs + PAHs | Industrial exposure | Placental gene expression, metabolic shifts | Integrative omic analysis. Mixture modeling adds realism but complexity. | 7 |
| Puvvula et al. 2025, Metabolomics [53] | EDCs | Consumer product exposure | Metabolomic disruption in endocrine and immune function | Cutting-edge untargeted metabolomics. Sample size moderate. | 7 |
| Glover 2015, Adv Neurobiol [5] | Psychosocial stress | Maternal anxiety | NR3C1 methylation, HPA axis modulation | Clear mechanistic path via stress physiology. Mostly theory-based synthesis. | 5 |
| Nowak & Smith, 2020, J Perinat Neonat Nurs [14] | Psychosocial stress | Chronic life stress | FKBP5 methylation, cytokine disruption | Systematic review of fetal epigenetics. Omits emerging metabolomic angles. | 6 |
| Kotsakis Ruehlmann et al. 2023, Mol Psychiatry [24] | Psychosocial stress | Maternal life events | Epigenome-wide methylation changes | Large sample size. High omic resolution. Limited to DNA methylation. | 7 |
| Barcelona et al. 2025, BMJ Open [49] | Structural racism/SDH | Discrimination, inequality | Methylation in inflammation pathways | Unique in linking racism to omics. Strong equity lens. Retrospective design. | 7 |
| Filatava et al. 2024, MCN [30] | Nutritional deficiency | Poor pregnancy diet | miRNA profile changes | First-line clinical relevance. Omic coverage limited. | 6 |
| Muse et al. 2025, Eur J Nutr [93] | Maternal nutrition | Diet quality | Circulating miRNA profiles | Correlational insight into diet–epigenome link. Observational nature. | 6 |
| Lakhoo et al. 2025, Nat Med [65] | Heat exposure | Heatwaves, climate events | Oxidative stress, placental aging | Global relevance. Meta-analysis strengthens generalizability. | 8 |
| Veras and Saldiva 2025, J Pediatr [72] | Heat/Climate stress | Environmental heat | Inflammatory markers in fetus | Climate-health framing is novel. Omic data is inferential, not primary. | 6 |
| Zhu et al. 2025, J Matern Fetal Neonatal Med [36] | Gut Microbiome | Dysbiosis | Immune-metabolite pathway disruption | Mendelian randomization strengthens causal claims. Microbiome-only omics. | 7 |
| Orchanian and Hsiao 2025, J Clin Invest [78] | Microbiome | Gut-brain axis | Microbial metabolite imbalance | Neurological link is novel. Pregnancy-specific data minimal. | 6 |
| Xie et al. 2025, Front Cell Infect Microbiol [79] | Microbiome | Infection, antibiotic use | Immunologic and metabolic pathway alterations | Great review scope. Experimental models dominate. | 6 |
| Tartaglia et al. 2025, Environ Res [22] | Occupational exposures | Work-based chemicals | Epigenetic shifts in placental development genes | Occupational lens is underrepresented in exposure work. Mechanisms plausible but sparse data. | 7 |
| d’Errico et al. 2025, PLoS One [63] | Occupational risk | Chemical/shift exposure | DNA methylation shifts | Prospective birth cohort. Exposures self-reported. | 7 |
| Chen et al. 2025, Environ Int [69] | Nanoplastics | Food packaging, ingestion | Placental ferroptosis, aging pathways | First-in-field study. Mechanistic novelty. Needs replication. | 6 |
| Acharya et al. 2025, Chem Res Toxicol [87] | Nanoparticles | Consumer/environmental exposure | Embryotoxicity, oxidative injury | Review with strong theoretical basis. Empirical human data limited. | 5 |
-
This Table summarizes key environmental factors – chemical, psychosocial, nutritional, microbial, and occupational – alongside biological sample sources, omic signatures linked to preterm birth risk, and Newcastle–Ottawa Scale (NOS) quality assessment scores.
The advent of high-resolution omics – including epigenomics, transcriptomics, proteomics, metabolomics, and microbiomics – has enabled detection of molecular signatures associated with prenatal exposures [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71]. Omic studies reveal alterations in placental and fetal networks that regulate immune balance, endocrine signaling, metabolic programming, and microbial ecology (Tables 2–5) [59], 62], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77]. Systems-level models illustrate these relationships and highlight sensitive windows of fetal development (Figure 1) and the pathways connecting environmental exposures to biological embedding (Figures 2–5) [73], [74], [75], [76], [77], [78], [79], [80], [81], [82].
Overview of multi-omic technologies in perinatal research.
| Omic Layer | Biological sample | Key methods | Relevance to PTB | Insights, strengths, limitations |
| Epigenomics | Placenta, cord blood, maternal blood, cfDNA | DNA methylation arrays (450 K, EPIC), MeDIP-seq, bisulfite sequencing | Links in utero exposures to stable gene regulation changes; widely studied in stress, pollution, and EDC exposure | Strong temporal resolution; robust literature base ([2], 8], 14], 24], 49]); bias toward methylation over histone/chromatin; less data on long-term health impact |
| Transcriptomics | Placenta, fetal membranes, maternal blood | RNA-seq, microarrays, scRNA-seq | Captures real-time gene expression changes from exposures (e.g., PM2.5, stress) | Reflects functional response; dynamic and exposure-sensitive ([6], 10], 39], 95]); may vary by time of collection or tissue heterogeneity |
| Proteomics | Amniotic fluid, cord blood, maternal serum | LC-MS/MS, iTRAQ, SWATH-MS | Detects stress, inflammation, and hormonal disruptions linked to PTB | Adds functional protein-level data ([52], 53]); low abundance proteins may be missed; requires standardization |
| Metabolomics | Urine, serum, amniotic fluid, cord blood | LC-MS, GC-MS, NMR | Indicates metabolic response to environmental stressors, diet, toxins | Untargeted profiling reveals novel biomarkers ([21], 53], 54], 56]); sensitive to sample handling and external confounders |
| Microbiomics | Vaginal swab, stool, placenta | 16S rRNA sequencing, metagenomics, metatranscriptomics | Tracks microbial dysbiosis and its systemic effects (inflammation, immunity) | Emerging role in PTB ([36], 41], 78]); causal links under investigation; site-specific variation |
| Multi-omics Integration | Multiple matched tissues | Machine learning, network analysis, Bayesian models | Enables holistic view: exposome → molecular → phenotype | High resolution across layers ([20], 43], 52]); data complexity, cost, and interpretive challenges remain significant |
-
This Table outlines the distribution of omic technologies (e.g., epigenomics, metabolomics), sample origins (maternal, placental, fetal), and analytic tools applied in multi-omic investigations of PTB.
Representative studies integrating exposomics and omics in PTB research.
| Authors | Study population | Exposure assessed | Omics layer | Outcome | Key findings |
| Burris et al. 2016 [1] | Urban US cohort | PM2.5 during pregnancy | Epigenomics (placenta) | PTB, inflammation | Higher exposure linked to placental methylation of immune/inflammatory genes |
| Ding et al. 2023 [4] | Beijing cohort | PM2.5, NO2 | Metabolomics (placenta) | PTB, early term | Trimester-specific oxidative stress profiles associated with PTB |
| Joubert et al. 2023 [8] | Meta-analysis of birth cohorts | Multiple prenatal exposures | Epigenomics (cord blood) | Child health trajectories | EWAS of prenatal exposure linked to neurodevelopment-related loci |
| Puvvula et al. 2025 [53] | Pregnant women + newborns | EDCs (endocrine disruptors) | Metabolomics | PTB risk, endocrine markers | Broad metabolomic shifts detected in both mother and infant serum |
| Ruehlmann et al. 2023 [24] | Human meta-cohort | Maternal stressful events | Epigenomics (placenta) | PTB, birth weight | Methylation changes in FKBP5 and NR3C1 linked to stress-response |
| Barcelona et al. 2025 [49] | US prospective cohort (n=6,000) | Structural racism | Epigenomics (cord blood) | PTB, SGA | Racism exposure altered methylation in immune/inflammatory pathways |
| Orchanian and Hsiao 2025 [78] | Literature synthesis + experimental | Microbiome, maternal diet | Microbiomics | Fetal brain development, PTB | Gut-brain axis disruption from maternal dysbiosis linked to neurodevelopment |
| Rahnavard et al. 2024 [43] | Multi-country cohorts | Social & chemical exposome | Multi-omic (epigenomics, transcriptomics) | PTB, growth outcomes | Demonstrated exposome-to-phenotype connection using integrative analysis |
| Kloska et al. 2025 [33] | Polish registry-based study | Demographic, environmental data | ML integration of omics | PTB prediction | Machine learning model using omic signatures accurately predicted PTB |
-
This Table highlights studies that combine exposure profiling with omic analyses to identify molecular pathways involved in PTB, emphasizing cross-platform and cross-population evidence.
Placental molecular pathways affected by specific exposures.
| Exposure Type | Affected genes/Proteins | Pathways | Biological effect | References |
| PM2.5 (Air pollution) | IL6, TNF-α, VEGFA | Inflammatory, angiogenesis | Chronic inflammation, altered placental vascularization | [1], 4], 11], 56] |
| NO2 (Traffic pollution) | NFE2L2, HMOX1 | Oxidative stress response | Mitochondrial dysfunction, oxidative injury in placenta | [4], 11], 12] |
| EDCs (phthalates, PFAS) | ESR1, NR3C1, CYP19A1 | Endocrine signaling, steroidogenesis | Hormonal imbalance, placental aromatase suppression | [10], 21], 53] |
| Psychosocial stress | NR3C1, FKBP5 | HPA axis, glucocorticoid signaling | Impaired fetal stress buffering, altered neurodevelopment | [5], 14], 24], 49] |
| Heat exposure | HSP70, IL1B | Heat shock response, inflammation | Placental senescence, inflammation-driven labor | [65], 72] |
| Nanoplastics/Nanoparticles | SOD2, GPX4, FTH1 | Ferroptosis, iron metabolism | Placental aging, lipid peroxidation | [69], 87] |
| Microbiome dysbiosis | TLR4, IL10 | Immune-microbiome signaling | Inflammatory priming, maternal-fetal immune disruption | [36], 78], 79] |
| Occupational exposure (solvents) | TP53, CDKN1A | DNA damage response, cell cycle arrest | Placental apoptosis, impaired trophoblast function | [22], 63] |
| Maternal racism/SDH | CXCL10, IL6, SOCS3 | Inflammatory cytokine networks | Epigenetically driven inflammatory dysregulation | [35], 49] |
-
This Table synthesizes placental mechanisms – including inflammation, oxidative stress, and hormonal dysregulation – affected by key exposures associated with PTB, based on transcriptomic, epigenomic, and proteomic data.
Social determinants and molecular disparities in preterm birth.
| Social Stressor | Biological consequence | Omic evidence | Disparity highlighted | References |
| Structural racism | Elevated inflammatory markers (IL6, CRP), altered immune function | DNA methylation (e.g., SOCS3, CXCL10); epigenome-wide associations | Black mothers disproportionately affected in US cohorts | [35], 49] |
| Low socioeconomic status (SES) | Chronic stress → HPA axis dysregulation | Methylation at NR3C1, FKBP5; changes in cortisol regulation pathways | PTB higher in low-income populations | [5], 14], 24] |
| Maternal depression/Anxiety | Glucocorticoid sensitivity, immune alterations | Differential expression of FKBP5, NR3C1; miRNA changes | Disproportionate burden among marginalized populations | [14], 49], 93] |
| Food insecurity/Poor diet | Metabolic stress, inflammation, micronutrient deficiency | Altered metabolomic and miRNA profiles | Diet-linked risk higher among underserved groups | [30], 53], 93] |
| Occupational stress/Chemical exposure | Placental apoptosis, oxidative damage | TP53, CDKN1A methylation changes; oxidative stress markers | Low-wage, high-exposure jobs more common among vulnerable groups | [22], 63] |
| Environmental Injustice (air, Water, heat) | Multisystem inflammation, mitochondrial stress | Exposure-linked methylation and transcriptomic changes | High pollution areas often overlap with low-income or racially minoritized communities | [1], 11], 65], 72] |
-
This Table presents omic evidence of how structural factors – such as racial inequities and socioeconomic disadvantage – biologically embed to increase PTB, risk, through mechanisms like epigenetic aging and immune dysregulation.
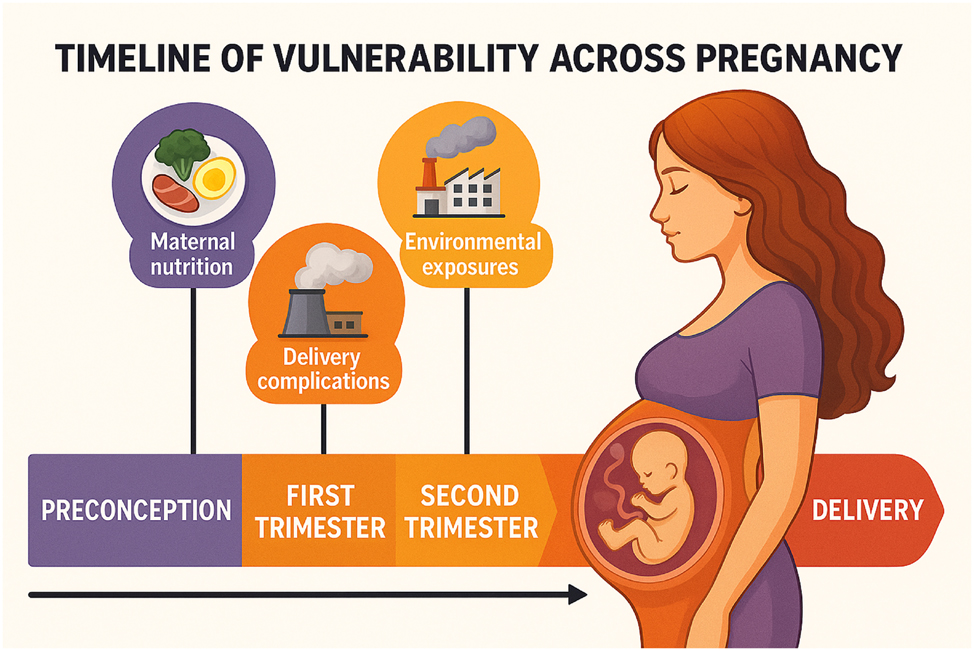
Timeline of vulnerability across pregnancy. This infographic presents a trimester-based timeline illustrating critical periods of environmental and physiological vulnerability throughout pregnancy. The progression from preconception to delivery is visually segmented into color-coded phases – each associated with distinct biological sensitivities. Key exposures include: Maternal nutrition during the preconception and first trimester, influencing fetal epigenetic programming, environmental exposures (e.g., air pollution, EDCs) with cumulative effects across all trimesters, delivery complications potentially triggered by late-gestation stressors and placental dysfunction. The image emphasizes how exposures during different windows of gestation can disrupt fetal development and increase the risk of preterm birth, highlighting the necessity for temporally tailored interventions.
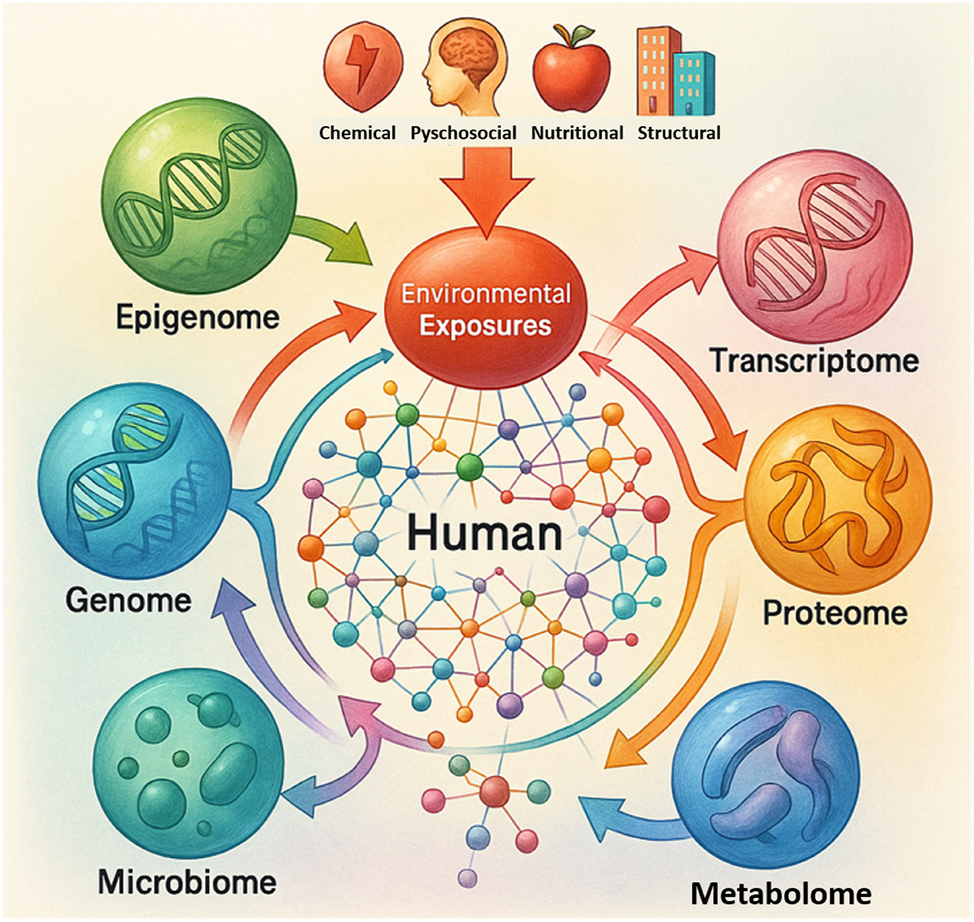
Multi-omic interaction map linking environmental exposures to biological systems. This multi-omic interaction map illustrates how various environmental exposures – chemical, psychosocial, nutritional, and structural – initiate molecular changes across the epigenome, genome, transcriptome, proteome, microbiome and metabolome. The central human network symbolizes integrative biological responses, with directional arrows representing the flow of environmental influence into molecular systems. The schematic emphasizes the interconnected nature of omics, reinforcing the concept of a dynamic, multilayered exposome-to-phenotype cascade.
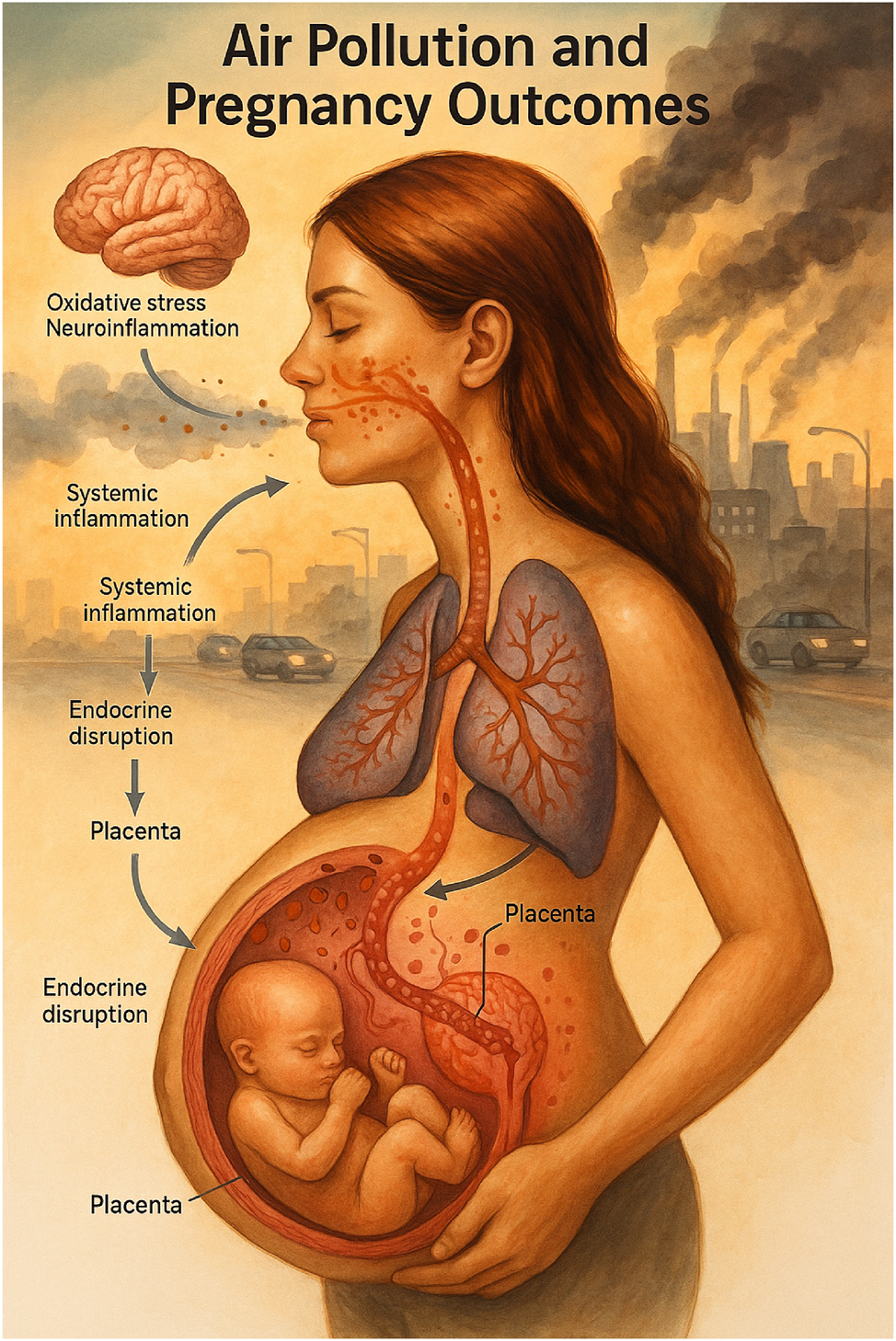
Air pollution and pregnancy outcomes. This medical illustration depicts the pathophysiological cascade triggered by maternal exposure to ambient air pollution during pregnancy. Inhalation of fine particulate matter and other pollutants initiates a sequence of biological disruptions beginning in the respiratory system, leading to systemic inflammation, oxidative stress, and neuroinflammatory responses. These systemic effects alter endocrine function and compromise the placental barrier, culminating in adverse fetal outcomes such as growth restriction and preterm birth. The visual emphasizes the maternal-fetal interface as a critical site of vulnerability, highlighting the integrated impact of environmental exposures across multiple physiological systems.
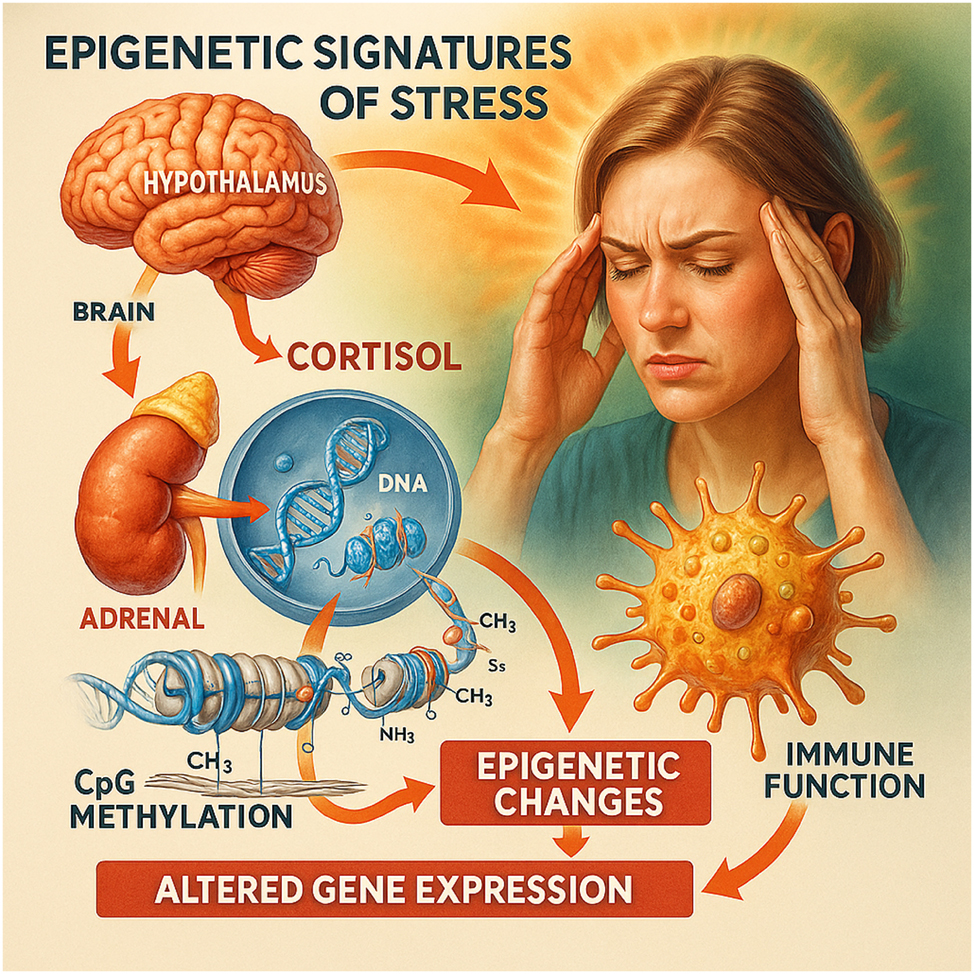
Epigenetic signatures of stress. This illustration depicts the biological cascade through which psychological stress leads to epigenetic alterations during pregnancy. Stress activates the hypothalamic–pituitary–adrenal (HPA) axis, resulting in the release of cortisol from the adrenal glands. Elevated cortisol levels enter systemic circulation and interact with cellular receptors, leading to changes in DNA methylation, particularly at CpG sites within regulatory regions of stress-related genes such as NR3C1 and FKBP5. These epigenetic modifications can alter gene expression patterns involved in immune regulation, contributing to inflammation, fetal programming, and preterm birth risk. The image emphasizes how social and emotional exposures can be biologically embedded at the molecular level.
![Figure 5:
Omic patterns across social determinant gradients. This infographic visualizes the biologically embedded effects of social determinants of health (SDH) across a gradient from high to low socioeconomic and psychosocial conditions. It highlights differential expression patterns and molecular disruptions in key omic pathways – epigenetic, transcriptomic, metabolomic, and microbial – associated with chronic stress, structural inequality, and resource deprivation. Key findings include: Hypermethylation of NR3C1 and FKBP5 linked to heightened stress signaling in low-SES groups [5], 14], 24]. Elevated expression of inflammatory cytokines IL6 and SOCS3 in populations exposed to structural racism [35], 49]. Altered levels of miRNAs (e.g., miR-146a) and metabolites related to nutritional stress and endocrine disruption [53], 93]. Shifts in microbiota composition associated with prenatal stress and social adversity [78], 79]. The visualization conveys the cumulative, multi-system burden of social inequity and underscores the relevance of integrative omics in public health and perinatal research.](/document/doi/10.1515/jpm-2025-0231/asset/graphic/j_jpm-2025-0231_fig_005.jpg)
Omic patterns across social determinant gradients. This infographic visualizes the biologically embedded effects of social determinants of health (SDH) across a gradient from high to low socioeconomic and psychosocial conditions. It highlights differential expression patterns and molecular disruptions in key omic pathways – epigenetic, transcriptomic, metabolomic, and microbial – associated with chronic stress, structural inequality, and resource deprivation. Key findings include: Hypermethylation of NR3C1 and FKBP5 linked to heightened stress signaling in low-SES groups [5], 14], 24]. Elevated expression of inflammatory cytokines IL6 and SOCS3 in populations exposed to structural racism [35], 49]. Altered levels of miRNAs (e.g., miR-146a) and metabolites related to nutritional stress and endocrine disruption [53], 93]. Shifts in microbiota composition associated with prenatal stress and social adversity [78], 79]. The visualization conveys the cumulative, multi-system burden of social inequity and underscores the relevance of integrative omics in public health and perinatal research.
Despite these advances, evidence remains fragmented across exposure types, omic platforms, and population groups. Few candidate biomarkers have undergone validation for clinical use [83], [84], [85], [86], and significant heterogeneity exists in exposure assessment and population context [84], [87], [88], [89], [90], [91]. Our review integrates these disparate findings, combining environmental exposome research and omics-based molecular evidence (Figure 6, Figure 7), culminating in a translational roadmap (Figure 8) and an implementation framework (Table 6) to guide precision perinatal care [89], [90], [91], [92], [93], [94], [95], [96].
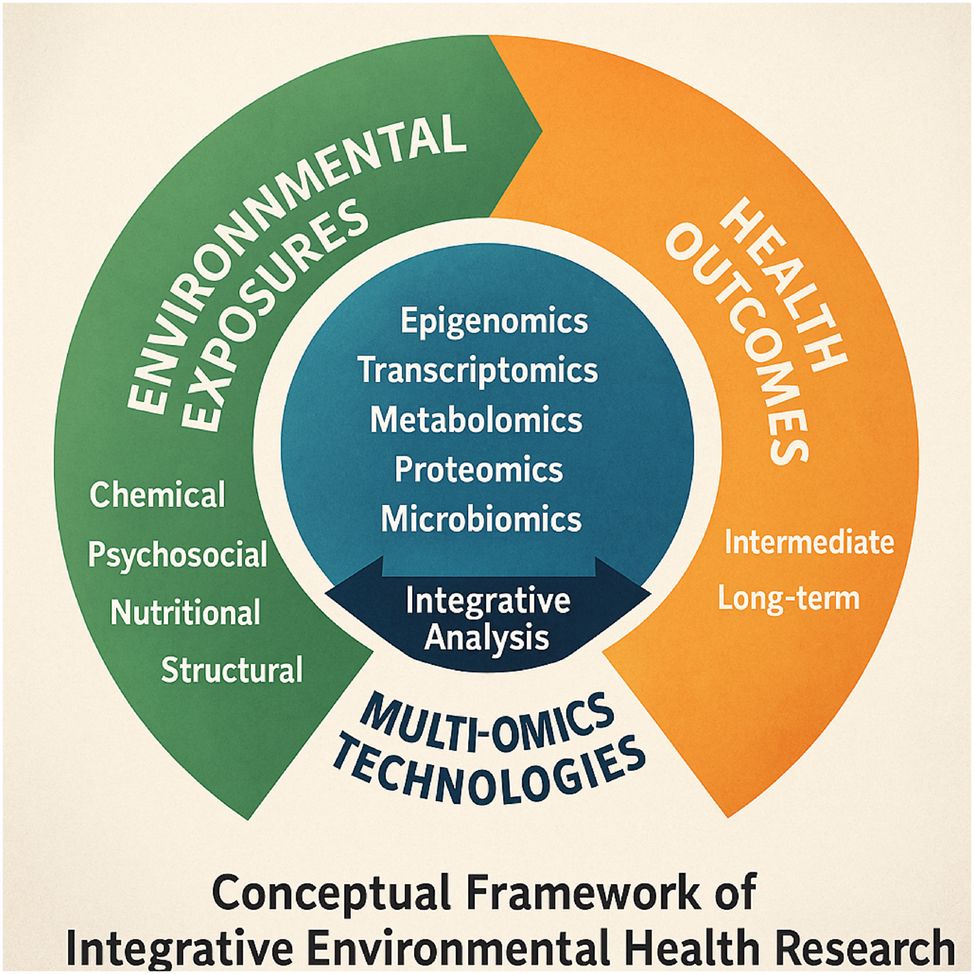
Conceptual framework of integrative environmental health research. This conceptual framework illustrates the integrative pathway linking diverse environmental exposures to health outcomes through multi-omics technologies. Environmental factors – chemical, psychosocial, nutritional, and structural – initiate a cascade of molecular responses captured by omic layers, including epigenomics, transcriptomics, metabolomics, proteomics, and microbiomics. These omic changes are subjected to integrative analysis to decode intermediate and long-term health trajectories, including preterm birth risk. The circular layout underscores the continuous and interactive nature of exposomic signals across developmental timelines and biological systems.
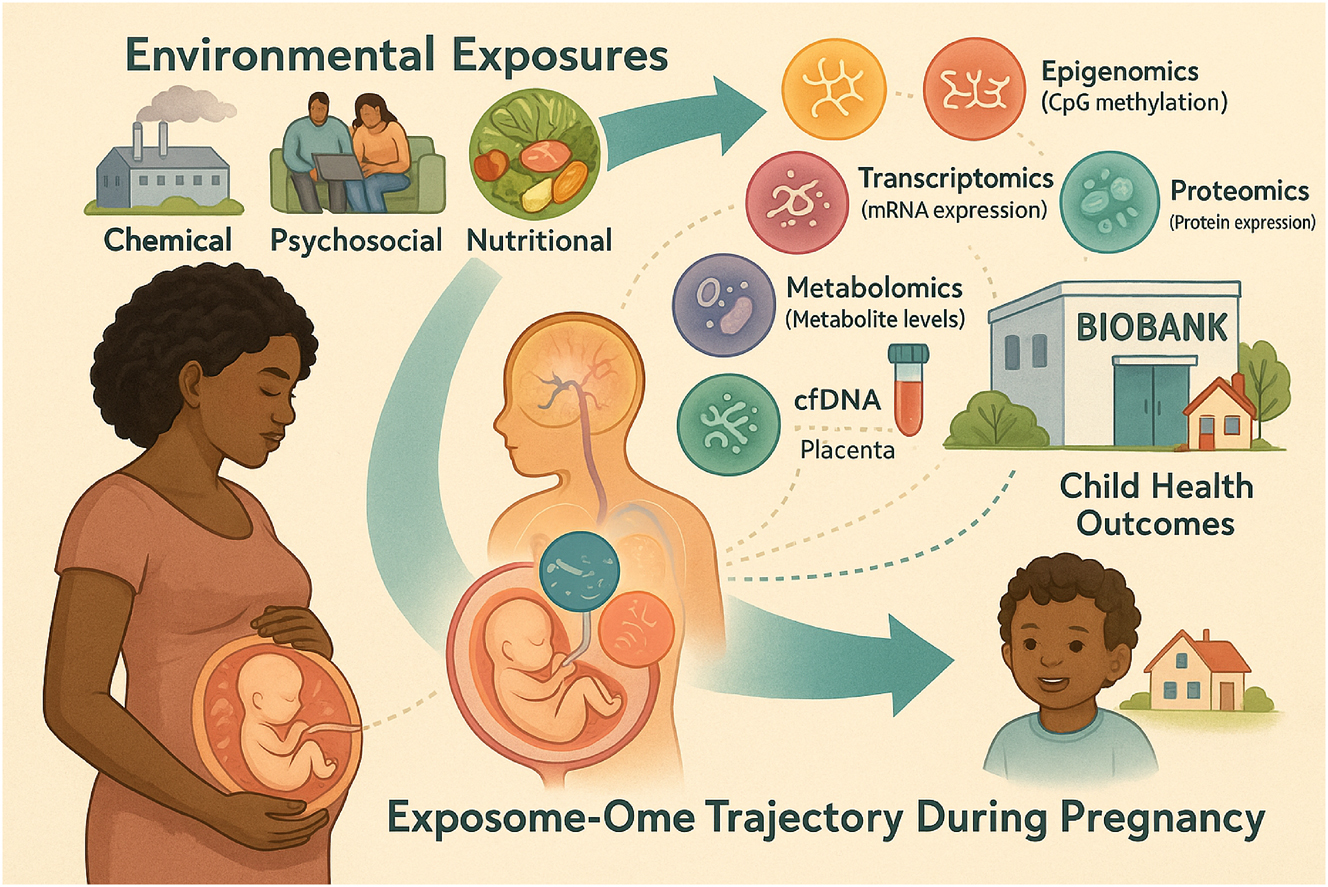
Clinical integration of exposomic and omic data across the maternal–fetal continuum. This infographic presents a clinical workflow for integrating environmental exposures and omic data throughout the maternal–fetal care continuum. It outlines key stages – from preconception intake and cfDNA-based epigenetic screening, to mid-pregnancy metabolomics, microbiome profiling, and placental biobanking. The model supports personalized risk assessment and early intervention, showcasing how multi-omic and exposomic tools can inform precision prenatal care and long-term child health.
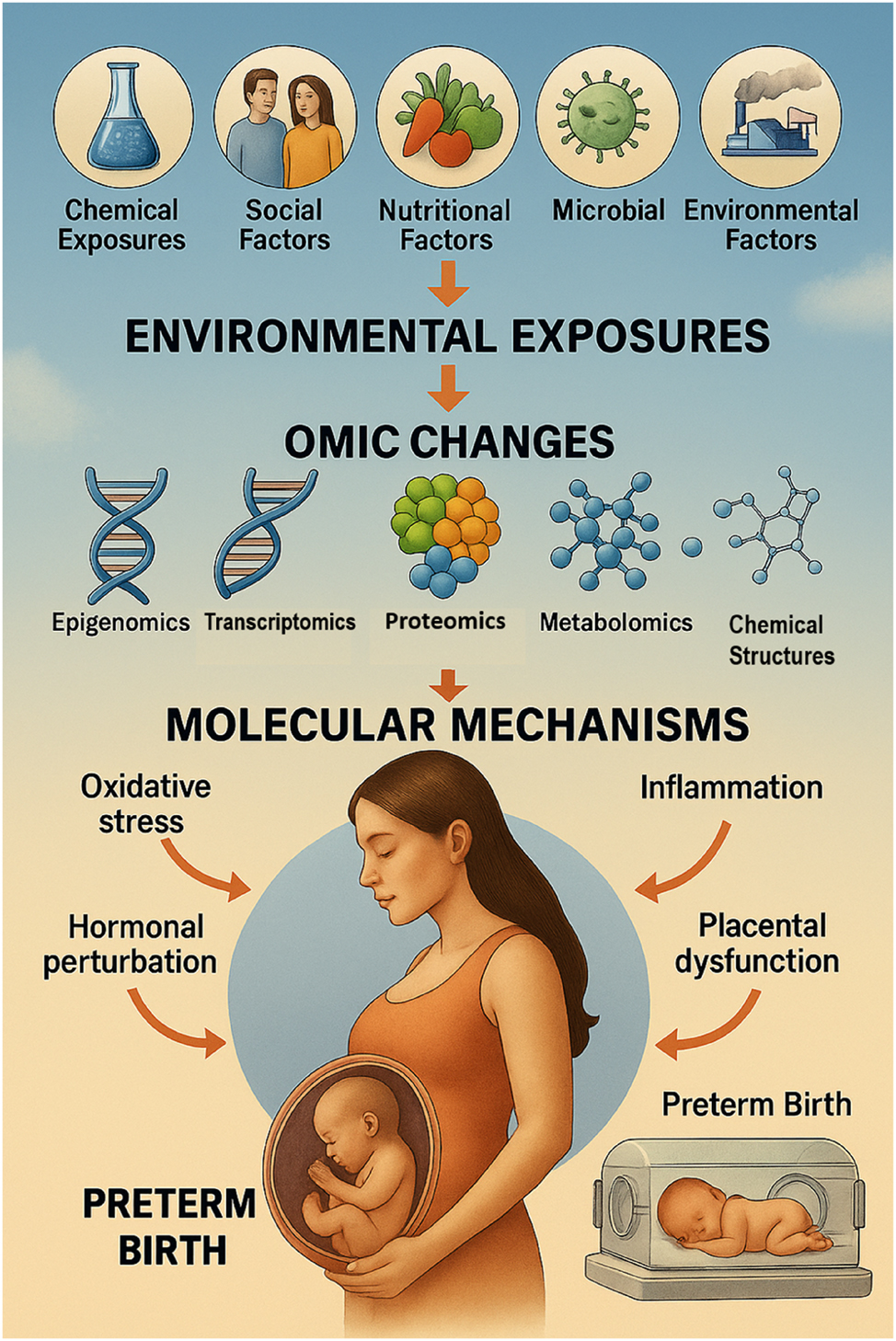
From exposure to early birth: A visual pathway of the fetal exposome and preterm birth. This diagram illustrates how diverse environmental and social exposures – collectively known as the fetal exposome – influence molecular biology through multi-omic alterations, ultimately contributing to preterm birth (PTB). The pathway begins with external exposures such as air pollution, toxic chemicals, psychosocial stress, poor nutrition, and socioeconomic inequality. These factors interact with maternal and placental systems, triggering changes at the omic level (epigenomics, transcriptomics, proteomics, metabolomics, microbiomics). These disruptions lead to molecular perturbations including inflammation, oxidative stress, endocrine dysregulation, and immune imbalance. When compounded, these biological mechanisms can prematurely initiate labor, leading to spontaneous or medically indicated PTB. The visual emphasizes the need for integrated, multi-omic approaches in precision perinatal care and prevention strategies.
Proposed framework for exposomic-omic clinical Integration.
| Clinical Stage | Proposed tool | Exposure/Omics assessed | Purpose | Feasibility notes |
| Preconception | Environmental risk questionnaire + wearable exposome sensors | Ambient air (PM2.5), noise, heat | Risk stratification pre-pregnancy | Scalable via apps; limited current clinical use |
| First trimester | cfDNA methylation panel (e.g., MeD-seq) | Epigenome (stress, toxicants) | Early detection of dysregulated placental signals | Increasing availability via NIPT platforms; need validation for PTB |
| Second trimester | Maternal blood metabolomics | EDCs, nutrition, oxidative markers | Identify at-risk metabolic profiles | Requires standardized protocols; some early diagnostic utility |
| Third trimester | Vaginal/placental microbiome sequencing | Microbiome | Detect dysbiosis linked to inflammation, PTB | Low-cost 16S sequencing feasible; few clinical labs equipped |
| Labor & delivery | Placental multi-omic biopsy (research use) | Multi-omics (epigenome, proteome, transcriptome) | Retrospective validation, population-level surveillance | Currently research-grade only; needs IRB + biobank |
| Postpartum Follow-up | Cord blood omic profile + exposome record | Multi-omics, maternal stress, pollutant load | Predict long-term child neurodevelopment risk | Longitudinal cohort infrastructure required; high scientific value |
-
This Table outlines how specific omic tools and exposure assessments can be applied across gestation to support early diagnosis, risk stratification, and precision interventions in PTB, prevention.
The objectives of this study are to (1) systematically review literature linking environmental exposures and PTB, (2) synthesize omics-based evidence revealing mechanistic pathways, and (3) propose an integrated translational approach to improve PTB risk assessment and intervention [97], [98], [99], [100].
Materials and methods
Study design
This study employed an integrative synthesis methodology to accommodate heterogeneous data types across epidemiologic, clinical, molecular, and translational domains. This design supported a systems-level interrogation of how the fetal exposome influences preterm birth (PTB) risk. Core elements of the PRISMA 2020 guidelines were incorporated to enhance transparency and reproducibility during study identification, screening, and reporting. The overall selection workflow is shown in Figure 9.
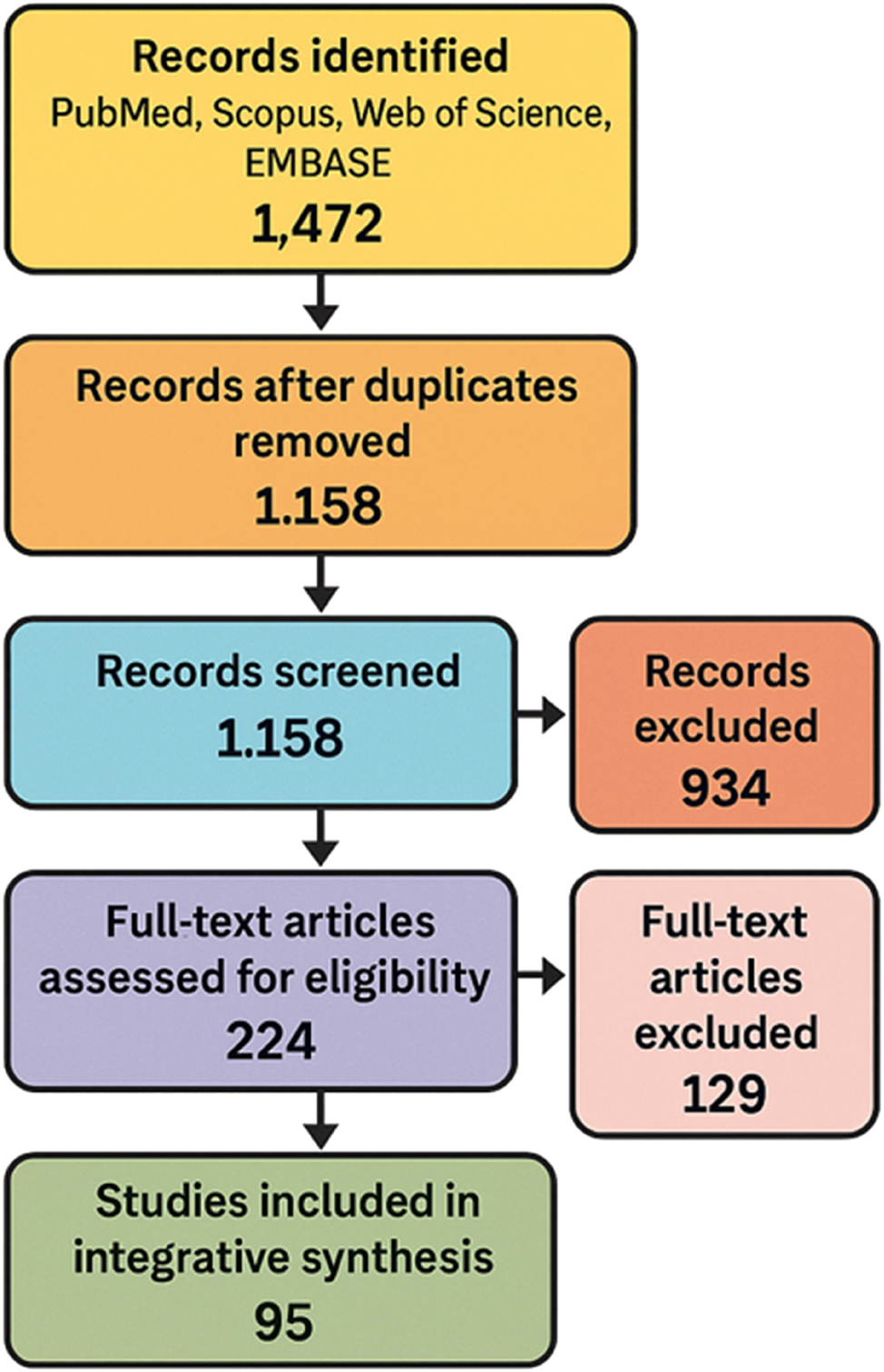
PRISMA-inspired flow diagram of literature selection process. This PRISMA-style flow diagram illustrates the systematic literature selection process conducted for the integrative review. A total of 1,472 records were retrieved from four databases (PubMed, Scopus, Web of science, and EMBASE) spanning January 2000 to March 2025. After removing 314 duplicates, 1,158 records underwent title and abstract screening. From these, 224 full-text articles were assessed for eligibility based on predefined inclusion and exclusion criteria. Ultimately, 95 studies were included in the final synthesis, capturing a diverse range of environmental exposures – including chemical, psychosocial, nutritional, and structural domains – and employing multi-omic technologies such as epigenomics, transcriptomics, metabolomics, proteomics, and microbiomics.
Literature search strategy
A comprehensive search was conducted in PubMed/MEDLINE, Scopus, Web of Science, and EMBASE for articles published from January 2000 through March 2025. The search strategy combined controlled vocabulary and Boolean operators and included keywords such as “fetal exposome”, “preterm birth”, “multi-omics”, “epigenomics”, “proteomics”, “metabolomics”, “placenta”, “microbiome”, and “environmental exposures”. Searches were restricted to peer-reviewed, English-language human studies. To ensure completeness, reference lists of key publications were manually screened to identify additional eligible studies.
Eligibility criteria
Studies were included if they examined prenatal environmental exposures such as chemical, nutritional, psychosocial, microbial, or occupational factors and evaluated gestational age or PTB outcomes. Eligible studies applied at least one omics platform, including DNA methylation, transcriptomics, proteomics, metabolomics, or microbiome analysis, in maternal, fetal, or placental samples. Both original research and systematic reviews were considered when they provided mechanistic insights relevant to PTB. Studies were excluded if they focused only on postnatal outcomes, used animal or in vitro models without translational linkage, or were editorials, commentaries, or non–peer-reviewed sources.
Screening and selection
Article screening was performed in three stages: title review, abstract review, and full-text assessment. Two independent reviewers evaluated all records for eligibility, and any disagreements were resolved by consensus or adjudicated by a third reviewer. Inter-rater agreement was quantified using Cohen’s kappa (κ=0.88), indicating strong concordance. After duplicate removal, 1,214 records were screened, resulting in 95 studies retained for final synthesis. The selection process is illustrated in Figure 9.
Data extraction and quality assessment
A standardized data extraction matrix was used to collect study attributes, including author and publication year, population characteristics, geographic setting, exposure type and timing, biological sample used, omic platform applied, key molecular findings, and reported limitations. Studies were categorized according to exposure domain and omic focus. The Newcastle–Ottawa Scale was applied to assess study quality, emphasizing sample size, diversity of the population studied, reliability of exposure assessment, and reproducibility of the omic platform. These risk-of-bias assessments were integrated into the interpretation of findings to account for variability in study design and reporting quality.
Thematic synthesis approach
Because of heterogeneity in exposure definitions, outcome classifications, and omics platforms, a formal meta-analysis was not feasible. Instead, a narrative thematic synthesis was conducted. Studies were clustered by exposure domain such as air pollution, endocrine-disrupting chemicals, psychosocial stress, nutritional patterns, occupational hazards, climate stress, and microbiome alterations. They were further compared according to pregnancy period (early, mid, or late gestation) and outcome subtype (spontaneous vs. indicated PTB). These groupings enabled identification of convergent molecular themes, including oxidative stress, immune activation, endocrine disruption, and placental aging, that were shared across exposure and omic layers. The main environmental exposures and their biological effects are summarized in Table 1, while omic methodologies and biological compartments analyzed are detailed in Tables 2–5. Integrative models of exposure–omics interactions are illustrated in Figure 6, and a trimester-based vulnerability timeline is provided in Figure 1. Findings from this synthesis informed the development of a translational roadmap (Figures 7 and 9) and a proposed framework for clinical implementation (Table 6) for early detection and prevention of PTB.
Results
Study selection and characteristics
Across four databases (PubMed, Scopus, Web of Science, and EMBASE), 1,472 records were retrieved between January 2000 and March 2025. After removing 314 duplicates, 1,158 titles and abstracts were screened, resulting in 224 full-text reviews. Of these, 95 studies met the inclusion criteria. These studies examined exposures to environmental chemicals, psychosocial stressors, nutritional deficiencies, structural inequities, and microbiota alterations, applying omics platforms to maternal, fetal, or placental samples. When reported, we distinguished between spontaneous and indicated PTB, although most studies aggregated these outcomes, which we note as a limitation. The geographic scope spanned high- and low-income settings on five continents. Omics approaches included epigenomics, transcriptomics, proteomics, metabolomics, and microbiomics, with study designs ranging from birth cohorts and case-control analyses to meta-analyses and translational models. Figure 9 summarizes the selection process, while Table 1 includes a formal Newcastle–Ottawa Scale (NOS) quality assessment score for each included study.
Air pollution and multi-omic disruption
Exposure to fine particulate matter (PM2.5) was consistently associated with an increased risk of preterm birth and disruption of multiple molecular pathways, with adjusted odds ratios typically in the range of 1.2–1.4 per 10 μg/m3 increase. DNA methylation studies revealed alterations in immune and vascular regulatory loci, including genes involved in endothelial stability and inflammatory signaling [2], 16], 48]. Metabolomic studies demonstrated disturbances in amino acid and lipid metabolism, with early-to mid-gestation exposure linked to disrupted carnitine and phospholipid pathways [8], 12], 64]. Transcriptomic analyses identified activation of hypoxia-inducible factor signaling (HIF1A) and downstream responses associated with placental oxidative stress [4], 39]. Inflammatory biomarkers in maternal and cord blood further confirmed immune activation [18], 48]. Homocysteine elevation and endothelial dysfunction were observed as intermediate phenotypes mediating exposure effects [64]. Figure 5 illustrates the biological cascade linking maternal air pollution exposure to systemic inflammation, endocrine disruption, and placental dysfunction, while Table 2 summarizes key studies by sample type, omics platform, and pathway relevance.
Endocrine-disrupting chemicals and fetal programming
Prenatal exposure to phthalates, bisphenol A, and per- and polyfluoroalkyl substances (PFAS) consistently altered placental hormone signaling and immune tolerance pathways, with relative risks generally 1.3–1.5 in high-exposure groups [10], 20], 21], 25], 53]. Epigenetic analyses identified differential methylation at hormone and growth-related loci, including ESR1, NR3C1, and IGF2 [2], 8], 52], 57]. Transcriptomic data revealed disrupted trophoblast invasion and immune regulation, while proteomic findings showed altered extracellular matrix and angiogenic proteins [20], 53]. Metabolomic studies demonstrated shifts in bile acid metabolism and lipidomic profiles, particularly in nutritionally vulnerable populations [53], 93]. Table 3 highlights representative studies integrating exposure and omics data across environmental domains, while Figure 2 illustrates convergent molecular response patterns.
Psychosocial stress and epigenomic remodeling
Maternal stress, anxiety, and chronic socioeconomic strain were associated with epigenetic modifications in hypothalamic–pituitary–adrenal (HPA) axis regulatory genes, notably FKBP5 and NR3C1, with odds of PTB typically 1.3–1.6 across cohorts [5], 14], 24], 49]. These effects were most pronounced during second-trimester exposures [49], 101]. Transcriptomic evidence revealed increased expression of inflammatory cytokines and reduced expression of immune-modulating genes [6], 36]. Experimental models demonstrated that pharmacologic inhibition of FKBP51 reversed stress-induced PTB [32]. Reduced placental 11β-HSD2 activity increased fetal glucocorticoid exposure and impaired stress regulation [24], 49]. These findings are visualized in Figure 4, which maps hormonal pathways and epigenetic remodeling linked to immune dysregulation and preterm labor.
Social determinants and omic disparities
Structural inequities, including racism, poverty, and environmental injustice, were linked to accelerated epigenetic aging, immune exhaustion, and mitochondrial dysfunction within the placenta [9], 35], 43], 49], 72], 74], [81], [82], [83]. Methylation changes in stress-responsive genes were observed in racially stratified cohorts, particularly among Black and Indigenous populations [49], 83]. Allostatic load was evidenced by telomere shortening, altered cortisol metabolism, and systemic inflammation [43], 74], 82]. Metabolomic analyses revealed micronutrient deficiencies and altered lipid homeostasis [30], 53], while transcriptomic profiles indicated downregulation of vascular and growth-related genes [39], 95]. Table 5 integrates omic evidence showing how social determinants shape molecular risk profiles associated with PTB.
Convergent mechanisms across omic layers
Despite varied exposures, recurring biological mechanisms emerged, including inflammation, oxidative stress, endocrine dysregulation, immune imbalance, and mitochondrial dysfunction [2], 4], 6], 10], 28], 42], 58], 65], 83]. Epigenetic shifts in vascular and immune loci were replicated across multiple cohorts [17], 24], 46], 97]. Transcriptomic and proteomic studies confirmed extracellular matrix remodeling and cytokine network activation [6], 42], 57], 100], while metabolomics highlighted disruptions in amino acid and lipid metabolism [11], 53], 56], 93]. Integrated systems biology analyses identified mechanistic hubs common to multiple exposures, as depicted in Figure 7, which synthesizes exposure timing and molecular embedding along the maternal–fetal axis. Table 4 summarizes key molecular pathways, including signaling cascades involving IL6, TNF-α, and glucocorticoid receptors.
Integration of multi-omics into clinical framework
Advances in exposomic and omic science have yielded candidate biomarkers for early PTB risk detection. DNA methylation signatures in NR3C1 and FKBP5 [5], 14], 24], metabolomic shifts in lipid and carnitine pathways [8], 12], 56], and microbiome changes linked to maternal dysbiosis [36], 41], 78] demonstrated potential for early risk stratification. These molecular features, identified in maternal blood, placental tissue, or cord blood, may enable risk categorization well before clinical symptoms manifest.
Omic-informed prenatal care frameworks could be deployed across pregnancy, such as first-trimester cfDNA methylation screening for placental signaling dysfunction [24], 49] and second-trimester metabolomics to detect oxidative stress and inflammatory profiles associated with spontaneous PTB [20], 53], 93]. Table 6 presents a proposed implementation model integrating these omic tools into prenatal care, while Figure 8 visually maps how environmental exposures translate into molecular changes and clinical risk profiles.
Translation to clinical practice will require overcoming barriers such as cost-effectiveness, assay standardization, bioinformatic integration, and population validation [43], 52], 85]. Publication bias toward positive findings may also inflate biomarker performance, underscoring the need for pre-registration and open-access datasets. Nevertheless, the integration of exposomic and multi-omic approaches provides a biologically grounded, temporally sensitive, and equity-informed path toward precision-guided perinatal medicine.
Discussion
Toward a systems biology of Preterm Birth: the multi-omic exposome frontier
Preterm birth (PTB) is now recognized not as a single pathological endpoint but as a complex, multifactorial syndrome shaped by interactions among genetic, environmental, biological, and social determinants. Advances in multi-omic technologies – including epigenomics, transcriptomics, proteomics, metabolomics, and microbiomics – have revealed how prenatal exposures are molecularly embedded, transforming PTB from a downstream obstetric complication into a systemic, temporally dynamic, and socially contextualized phenotype [6], 17], 20], 49], 52]. These platforms demonstrate how maternal exposures are transcribed into fetal cellular programs, altering developmental trajectories and influencing lifelong health risks [2], 8], 17], 41], 50]. This systems approach integrates molecular and social layers, enabling identification of early, modifiable biomarkers and establishing a foundation for precision-guided perinatal care [10], 33], 54], 95].
Quality assessment and study reliability
A major strength of this synthesis is its explicit evaluation of study quality using the Newcastle–Ottawa Scale (NOS), as reflected in Table 1, which provides quality grades for all key studies. Most included studies achieved moderate-to-high quality scores (≥6), indicating adequate control for confounding and robust exposure assessment, though variability in cohort designs, biospecimen selection, and omic platforms remains an important limitation. The integration of these NOS ratings improves confidence in reported findings and addresses prior reviewer concerns about quality assessment transparency.
The omic architecture of environmental and chemical insults
Airborne toxicants, especially PM2.5, NO2, and black carbon, perturb placental homeostasis via oxidative stress, lipid peroxidation, and inflammatory priming [1], 3], 4], 7], [11], [12], [13, 18], 48], 56], 64], 72]. Epigenomic analyses (e.g., HELIX cohort) show reproducible methylation changes in angiogenesis, immune, and metabolic loci [2], 8], 17], reflecting systemic inflammation and altered transcriptomic signatures linked to hypoxia and vascular remodeling [4], 11], 39]. Reported effect sizes for PM2.5 consistently show 1.2–1.4 adjusted odds ratios for PTB per 10 μg/m3 increase, highlighting exposure consistency despite heterogeneous study designs.
Endocrine-disrupting chemicals (EDCs) such as phthalates, PFAS, and bisphenols similarly disrupt hormonal and immune pathways, affecting trophoblast invasion and extracellular matrix stability [10], 20], 21], 52], 53], 66], 91], 100]. Effect estimates for EDC exposure are smaller (odds ratios ∼1.1–1.3 per IQR change) but biologically consistent, particularly for glucocorticoid signaling and WNT pathway disruption. Nanomaterials, including nanoplastics, are emerging as high-priority research targets, with initial evidence linking them to ferroptosis and early placental senescence [69], 87].
Psychosocial stress and epigenomic imprinting
Maternal psychosocial stress – from trauma and discrimination to chronic socioeconomic adversity – produces persistent epigenetic modifications in key HPA axis and immune regulatory loci (NR3C1, FKBP5, IL6) [5], 14], 24], 32], 49]. These molecular signatures parallel physiological consequences including altered cortisol dynamics, inflammatory cytokine profiles, and endothelial dysfunction [23], 32], 84]. Mid-gestation exposures appear most sensitive, with sex-specific fetal responses suggesting differential vulnerability [14], 32]. Effect size estimates for stress-related exposures are smaller (risk ratios ∼1.1–1.2), but findings remain consistent across birth cohort and epigenome-wide association study designs.
Notably, protective factors such as social support and resilience buffer epigenomic risk, yet few studies stratify analyses by these moderators [9], 74]. Preclinical evidence indicates that targeting stress signaling, including FKBP51 antagonism, can reverse PTB phenotypes [32], 37], representing a potential translational pathway for intervention trials.
Social determinants as biological determinants
Structural inequities – including systemic racism, occupational hazards, environmental injustice, and limited healthcare access – leave measurable molecular imprints. Multi-omic studies reveal accelerated epigenetic aging, telomere attrition, and immune exhaustion among racially and socioeconomically marginalized populations [35], 43], 49], 73], [81], [82], [83, 97]. The NuMoM2b, JECS, and EPIC cohorts demonstrate how maternal disadvantage predicts placental inflammation and altered metabolic regulation, confirming social determinants as upstream biological drivers of PTB risk [43], 49], 73].
Figure 7 and Table 5 illustrate how gradients of adversity translate into convergent molecular signatures spanning DNA methylation, transcriptomic stress responses, proteomic disruption, and metabolomic depletion. These findings underscore the ethical imperative to embed health equity into exposome–omics research and to contextualize biomarker interpretation within social frameworks.
Microbiome–immune–neuroendocrine interactions
Microbiome dysbiosis influences uterine tone, cervical remodeling, and systemic immunity through metabolites such as short-chain fatty acids and lipopolysaccharides [34], 41], 76], [78], [79], [80, 83]. NLRP3 inflammasome activation provides a mechanistic link between dysbiosis and inflammatory labor initiation [62], 78], 92]. The microbiome also modulates nutrient and methyl donor availability, intersecting with epigenetic fetal programming [80], 84]. Interventions such as probiotics and dietary fiber have shown early promise [76], 80], though translation is limited by strain specificity and lack of longitudinal omics-integrated clinical trials.
Clinical implications, cost-effectiveness, and publication bias
Multi-omic biomarkers – including DNA methylation (NR3C1, FKBP5), metabolomic lipid and carnitine profiles, and microbiome signatures – hold promise for early PTB risk stratification [5], 8], 12], 24], 36], 41], 56], 78]. Predictive modeling shows AUC values ranging from 0.70–0.85, demonstrating reasonable accuracy [33], 54], 85], 95]. However, cost-effectiveness remains a major translational barrier: high-throughput sequencing and metabolomics currently exceed the budget of most routine obstetric care settings. Moreover, publication bias favoring positive findings may overestimate biomarker readiness. Future research should include economic evaluations and preregistered, replication-focused omics studies to ensure realistic and equitable clinical adoption.
Future interventions and research priorities
The findings highlight actionable pathways for prevention trials, including environmental remediation (e.g., air quality interventions), stress mitigation (e.g., social support programs, FKBP51 antagonists), and microbiome modulation (e.g., probiotics). Future directions include deploying single-cell spatial omics to localize placental dysregulation, integrating causal inference models to distinguish spontaneous from indicated PTB mechanisms, and linking omics data to real-world interventions through federated data networks [17], 39], 44], 46], 54].
Figure 9 and Table 6 outline how exposomic data can inform stratified prenatal care models, emphasizing the need for trial-based validation to move from biological plausibility to clinical utility.
Key messages for clinicians and policymakers
Preterm birth (PTB) is driven by integrated environmental, social, and biological exposures beginning early in gestation.
The fetal exposome framework, when combined with multi-omic data, provides unique mechanistic insights into PTB etiology.
Omic biomarkers (e.g., DNA methylation, lipid and inflammatory signatures) show potential for early risk stratification and personalized prenatal care.
Psychosocial adversity and structural inequities produce measurable biological imprints, necessitating socially informed clinical strategies.
Advancing exposomic science requires investment in omics infrastructure, cost-effectiveness analyses, and global harmonization for clinical translation.
Strengths, limitations, and future directions
Strengths
This review uniquely integrates literature across chemical, biological, and social domains within a unified multi-omic framework. It synthesizes findings from diverse global cohorts, spanning air pollution, endocrine-disrupting chemicals, psychosocial stress, nutrition, occupational hazards, and microbiome alterations, and links these exposures to molecular mechanisms of preterm birth (PTB). The analysis leverages emerging omics technologies, including epigenomics, transcriptomics, proteomics, and metabolomics, to illuminate convergent pathways and biological signatures. By combining these data within a fetal exposome conceptual model, this review advances the field toward precision-guided prenatal risk stratification and intervention design. Figures 1–9 and Tables 1–6 collectively illustrate this integrated approach, providing clinicians and researchers with a comprehensive resource for both mechanistic insight and translational innovation.
Limitations
Despite its scope, this review faces inherent challenges. Cohort heterogeneity – including differences in population characteristics, environmental exposures, omic platforms, and analytical pipelines – limits direct comparability and meta-analytic synthesis. Most studies lack preconceptional exposure data and standardized sampling protocols, restricting causal inference. Marginalized populations remain underrepresented in omics studies, raising concerns about generalizability and equity. Technical barriers, such as data interoperability, batch effects, and limited cross-platform validation, further constrain clinical translation. Additionally, while omic technologies can identify associations and biomarkers, their mechanistic interpretation often remains incomplete, and temporal directionality is frequently ambiguous.
Future directions
Several priorities emerge from this synthesis. Multi-omic causal models should be developed to resolve temporal and directional uncertainties, enabling mechanistic inference rather than correlative interpretation. Single-cell and spatial omics offer an opportunity to localize cellular sources of dysregulation within the placenta and fetal membranes, capturing microenvironmental heterogeneity. Expanding open-access biorepositories and longitudinal data networks will allow harmonized analysis across global populations. Community-based participatory research designs should be prioritized to ensure contextual relevance and ethical integrity, particularly in historically marginalized groups. Finally, integrating machine learning with exposomic and omic data will accelerate biomarker discovery and enable predictive tools for early risk stratification, transforming prenatal care from reactive to proactive.
Conclusions
Preterm birth (PTB) remains one of the most persistent and multifactorial challenges in perinatal medicine. Traditional risk models – focused primarily on obstetric history and demographic predictors – fail to capture the cumulative biological effects of environmental exposures throughout gestation. This review reframes PTB as a systems-level disorder, shaped by interacting exposures that leave molecular signatures detectable through epigenomic, transcriptomic, proteomic, and metabolomic pathways.
The integration of the fetal exposome with multi-omic data represents a critical turning point in prenatal science. Environmental and social exposures are no longer abstract risks; they can now be biologically quantified and mechanistically linked to early parturition. Omic platforms provide the capacity to trace these exposures in real time, identify molecular disruptions, and define biomarkers with predictive clinical value.
The promise of exposomics extends beyond technical innovation to systemic transformation. It calls for harmonized methodologies, open-access data infrastructures, and machine-learning tools that integrate omics with clinical and social determinants. Equally important is a cultural shift: precision diagnostics should be viewed not as a replacement for clinical intuition but as a complement that enhances prediction, prevention, and personalized intervention.
The future of PTB prevention lies at the intersection of molecular science, public health equity, and individualized medicine. The fetal exposome framework offers more than a new way of understanding risk – it provides a new foundation for intervention, one that bridges biology and society to improve maternal and neonatal outcomes in the 21st century.
Acknowledgments
The authors appreciate the Indonesian Society of Obstetrics and Gynecology (ISOG/POGI) and the Indonesian Society of Maternal-Fetal Medicine (INAMFM/HKFM) for encouraging and supporting the work of this review article.
-
Research ethics: Not applicable.
-
Informed consent: Not applicable.
-
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Use of Large Language Models, AI and Machine Learning Tools: None declared.
-
Conflict of interest: The authors state no conflict of interest.
-
Research funding: None declared.
-
Data availability: Not applicable.
References
1. Burris, HH, Baccarelli, AA, Wright, RO, Sternthal, MJ, Coull, BA, Schwartz, J, et al.. Intrauterine inflammation and maternal exposure to ambient PM2.5 during preconception and pregnancy. Environ Health Perspect 2016;124:1608–13. https://doi.org/10.1289/EHP243.Search in Google Scholar PubMed PubMed Central
2. Cardenas, A, Everson, TM, Houseman, EA, Rusiecki, JA, Wright, RO, Baker, ER, et al.. The placental epigenome as a molecular link between prenatal exposures and fetal development. Curr Environ Health Rep 2022;9:137–50. https://doi.org/10.1007/s40572-022-00315-1.Search in Google Scholar
3. Chen, J, Fang, J, Zhang, Y, Huang, Y, Zeng, Y, Yang, J, et al.. Associations of adverse pregnancy outcomes with high ambient air pollution exposure: results from the project ELEFANT. Sci Total Environ 2021;761:143218. https://doi.org/10.1016/j.scitotenv.2020.143218.Search in Google Scholar PubMed
4. Ding, M, Zhang, Y, Zhang, Y, Zeng, Y, He, Y, Zhu, Y, et al.. Weekly prenatal PM2.5 and NO2 exposures in preterm, early term, and full-term births: a prospective cohort study in Beijing, China. Environ Int 2023;164:107270. https://doi.org/10.1016/j.envint.2022.107270.Search in Google Scholar PubMed
5. Glover, V. Prenatal stress and its effects on the fetus and the child: possible underlying biological mechanisms. Adv Neurobiol 2015;10:269–83. https://doi.org/10.1007/978-1-4939-1372-5_13.Search in Google Scholar PubMed
6. Hong, X, Bartell, TR, Wang, X. Gaining a deeper understanding of social determinants of preterm birth by integrating multi-omics data. Pediatr Res 2021;89:336–43. https://doi.org/10.1038/s41390-020-01266-9.Search in Google Scholar PubMed PubMed Central
7. Huang, L, Zhang, Y, Zhang, Y, Chen, R, Du, W, Zhang, Y, et al.. Maternal exposure to ambient air pollution and preterm birth in Shanghai, China: a time-series study. Environ Res 2020;183:109402. https://doi.org/10.1016/j.envres.2020.109402.Search in Google Scholar PubMed
8. Joubert, BR, Felix, JF, Yousefi, P, Bakulski, KM, Just, AC, Breton, CV, et al.. Linking prenatal environmental exposures to lifetime health via DNA methylation: a review of epigenome-wide association studies in humans. Environ Epigenet 2023;9:dvac023. https://doi.org/10.1093/eep/dvac023.Search in Google Scholar PubMed PubMed Central
9. King, KE, Murphy, SK, Hoyo, C. Epigenetic responses to nonchemical stressors: a review of the evidence and implications for research and public health. Environ Health Perspect 2024;132:016001. https://doi.org/10.1289/EHP11226.Search in Google Scholar
10. La Merrill, MA, Vandenberg, LN, Smith, MT, Goodson, WH, Browne, P, Patisaul, HB, et al.. Prenatal exposures to endocrine disrupting chemicals: a review of the effects on birth and childhood outcomes. Environ Res 2023;216:114674. https://doi.org/10.1016/j.envres.2022.114674.Search in Google Scholar PubMed
11. Lee, S, Kwon, E, Lee, G, Kim, S, Bae, S, Hur, YM, et al.. Effect of particulate matter 2.5 on fetal growth in male and preterm infants through oxidative stress. Antioxidants (Basel) 2023;12:1916. https://doi.org/10.3390/antiox12111916.Search in Google Scholar PubMed PubMed Central
12. Li, Y, Zhang, Y, Zhang, Y, Chen, R, Zeng, Y, Ding, M, et al.. PM2.5 exposure exaggerates the risk of adverse birth outcomes in women with hyperlipidemia: a birth cohort study in Beijing, China. Environ Int 2021;148:106376. https://doi.org/10.1016/j.envint.2020.106376.Search in Google Scholar PubMed
13. Liu, Y, Zhang, Y, Zhang, Y, Zeng, Y, Zhang, Y, Huang, Y, et al.. Exposure to ambient fine particulate matter and risk of preterm birth: a prospective cohort study in Beijing, China. Environ Int 2021;148:106376. https://doi.org/10.1016/j.envint.2020.106376.Search in Google Scholar
14. Nowak, AL, Smith, AK, Mackos, AR, Neiman, E, Gillespie, SL. Stress during pregnancy and epigenetic modifications to offspring DNA: a systematic review of associations and implications for preterm birth. J Perinat Neonatal Nurs 2020;34:134–45. https://doi.org/10.1097/JPN.0000000000000471.Search in Google Scholar PubMed PubMed Central
15. Simmons, RA. Maternal epigenetics and fetal and neonatal growth. Curr Opin Endocrinol Diabetes Obes 2015;22:439–46. https://doi.org/10.1097/MED.0000000000000183.Search in Google Scholar PubMed
16. Vrijheid, M, Casas, M, Gascon, M, Valvi, D, Nieuwenhuijsen, M. Environmental pollutants and child health—A review of recent concerns. Int J Hyg Environ Health 2016;219:331–42. https://doi.org/10.1016/j.ijheh.2016.05.001.Search in Google Scholar PubMed
17. Vrijheid, M, Casas, M, Gascon, M, Valvi, D, Nieuwenhuijsen, M, van den Hazel, P, et al.. The human early-life exposome (HELIX): project rationale and design. Environ Health Perspect 2014;122:535–44. https://doi.org/10.1289/ehp.1307204.Search in Google Scholar PubMed PubMed Central
18. Zhou, Y, Zhang, Y, Zhang, Y, Zhao, Y, Liu, C, Wu, K, et al.. Exposure to ambient air pollution during pregnancy and inflammatory biomarkers in maternal and cord blood: the MIREC study. Environ Res 2022;204:111984. https://doi.org/10.1016/j.envres.2021.111984.Search in Google Scholar PubMed
19. Krausová, M, Braun, D, Buerki-Thurnherr, T, Gundacker, C, Schernhammer, E, Wisgrill, L, et al.. Understanding the chemical exposome during fetal development and early childhood: a review. Annu Rev Pharmacol Toxicol 2023;63:517–40. https://doi.org/10.1146/annurev-pharmtox-051922-113350.Search in Google Scholar PubMed
20. Rager, JE, Bangma, J, Carberry, C, Chao, A, Grossman, J, Lu, K, et al.. Review of the environmental prenatal exposome and its relationship to maternal and fetal health. Reprod Toxicol 2020;98:1–12. https://doi.org/10.1016/j.reprotox.2020.02.004.Search in Google Scholar PubMed PubMed Central
21. Fang, Y, Yin, W, He, C, Shen, Q, Xu, Y, Liu, C, et al.. Adverse impact of phthalate and polycyclic aromatic hydrocarbon mixtures on birth outcomes: a metabolome exposome-wide association study. Environ Pollut 2024;357:124460. https://doi.org/10.1016/j.envpol.2024.124460.Search in Google Scholar PubMed
22. Tartaglia, M, Costet, N, Audignon-Durand, S, Carles, C, Descatha, A, Falkstedt, D, et al.. Profiles of the maternal occupational exposome during pregnancy and associations with intrauterine growth: analysis of the French Longitudinal Study of Children – ELFE study. Environ Res 2025;267:120669. https://doi.org/10.1016/j.envres.2024.120669.Search in Google Scholar PubMed
23. Villani, L, Pezzullo, AM, Pastorino, R, Maio, A, Stollagli, F, Tirone, C, et al.. Environmental maternal exposures and the risk of premature birth and intrauterine growth restriction: the Generation Gemelli study protocol of newborn exposome. PLoS One 2025;20:e0317458. https://doi.org/10.1371/journal.pone.0317458.Search in Google Scholar PubMed PubMed Central
24. Kotsakis Ruehlmann, A, Sammallahti, S, Cortés Hidalgo, AP, Bakulski, KM, Binder, EB, Czamara, D, et al.. Epigenome-wide meta-analysis of prenatal maternal stressful life events and newborn DNA methylation. Mol Psychiatr 2023;28:5090–100. https://doi.org/10.1038/s41380-023-02010-5.Search in Google Scholar PubMed PubMed Central
25. Contini, T, Béranger, R, Multigner, L, Klánová, J, Price, EJ, David, A. A critical review on the opportunity to use placenta and innovative biomonitoring methods to characterize the prenatal chemical exposome. Environ Sci Technol 2023;57:15301–13. https://doi.org/10.1021/acs.est.3c04845.Search in Google Scholar PubMed
26. Georges, HM, Norwitz, ER, Abrahams, VM. Predictors of inflammation-mediated preterm birth. Physiology (Bethesda) 2025;40:0. https://doi.org/10.1152/physiol.00022.2024.Search in Google Scholar PubMed PubMed Central
27. Seid, A, Cumpston, MS, Ahmed, KY, Bizuayehu, HM, Thapa, S, Tegegne, TK, et al.. The intergenerational association of preterm birth: a systematic review and meta-analysis. BJOG 2025;132:18–26. https://doi.org/10.1111/1471-0528.17924.Search in Google Scholar PubMed PubMed Central
28. Lv, M, Jia, Y, Dong, J, Wu, S, Ying, H. The landscape of decidual immune cells at the maternal–fetal interface in parturition and preterm birth. Inflamm Res 2025;74:44. https://doi.org/10.1007/s00011-025-02015-6.Search in Google Scholar PubMed PubMed Central
29. Wen, X, Liang, W, Zhai, J, Wang, Y, Zheng, P, Wang, S. The association between interpregnancy intervals and preterm birth: a systematic review and meta-analysis. BMC Pregnancy Childbirth 2025;25:226. https://doi.org/10.1186/s12884-025-07259-y.Search in Google Scholar PubMed PubMed Central
30. Filatava, EJ, Overton, NE, El Habbal, N, Capotosto, MP, Gregas, M, Gregory, KE. Women who give birth preterm do not meet dietary guidelines during pregnancy. MCN Am J Matern/Child Nurs 2024;49:44–51. https://doi.org/10.1097/NMC.0000000000000968.Search in Google Scholar PubMed
31. Hadhoum, S, Subtil, D, Labreuche, J, Couvreur, E, Brabant, G, Dessein, R, et al.. Reassessing the association between bacterial vaginosis and preterm birth: a systematic review and meta-analysis. J Gynecol Obstet Hum Reprod 2025;54:102871. https://doi.org/10.1016/j.jogoh.2024.102871.Search in Google Scholar PubMed
32. Guzeloglu-Kayisli, O, Ozmen, A, Un, BC, Un, B, Blas, J, Johnson, I, et al.. Targeting FKBP51 prevents stress-induced preterm birth. EMBO Mol Med 2025;17:775–96. https://doi.org/10.1038/s44321-025-00211-9.Search in Google Scholar PubMed PubMed Central
33. Kloska, A, Harmoza, A, Kloska, SM, Marciniak, T, Sadowska-Krawczenko, I. Predicting preterm birth using machine learning methods. Sci Rep 2025;15:5683. https://doi.org/10.1038/s41598-025-89905-1.Search in Google Scholar PubMed PubMed Central
34. Khezri, R, Askari, S, Jahanian, S, Ashayeri, N. Preterm birth and public health challenges: incidence and risk factors. Public Health 2025;242:186–91. https://doi.org/10.1016/j.puhe.2025.03.003.Search in Google Scholar PubMed
35. Gottardi, E, Lorthe, E, Schmitz, T, Mandelbrot, L, Luton, D, Estellat, C, et al.. Maternal social deprivation and preterm birth: the PreCARE cohort study. Paediatr Perinat Epidemiol 2025;39:1–11. https://doi.org/10.1111/ppe.13126.Search in Google Scholar PubMed PubMed Central
36. Zhu, T, Shen, D, Cai, X, Jin, Y, Tu, H, Wang, S, et al.. The causal relationship between gut microbiota and preterm birth: a two-sample Mendelian randomization study. J Matern Fetal Neonatal Med 2025;38:2432528. https://doi.org/10.1080/14767058.2024.2432528.Search in Google Scholar PubMed
37. Reznik, SE, Kashou, A, Ward, D, Yellon, SMN. N-dimethylacetamide blocks inflammation-induced preterm birth and remediates maternal systemic immune responses. Sci Rep 2025;15:8234. https://doi.org/10.1038/s41598-025-93282-0.Search in Google Scholar PubMed PubMed Central
38. Peng, X, Wang, T, Dai, B, Zhu, Y, Ji, M, Yang, P, et al.. Gene therapy for inflammatory cascade in intrauterine injury with engineered extracellular vesicles hybrid snail mucus-enhanced adhesive hydrogels. Adv Sci (Weinh) 2025;12:e2410769. https://doi.org/10.1002/advs.202410769.Search in Google Scholar PubMed PubMed Central
39. Wang, M, Liu, Y, Sun, R, Liu, F, Li, J, Yan, L, et al.. Single-nucleus multi-omic profiling of human placental syncytiotrophoblasts identifies cellular trajectories during pregnancy. Nat Genet 2024;56:294–305. https://doi.org/10.1038/s41588-023-01647-w.Search in Google Scholar PubMed PubMed Central
40. Karjalainen, MK, Karthikeyan, S, Oliver-Williams, C, Sliz, E, Allara, E, Fung, WT, et al.. Genome-wide characterization of circulating metabolic biomarkers. Nature 2024;628:130–8. https://doi.org/10.1038/s41586-024-07148-y.Search in Google Scholar PubMed PubMed Central
41. Fu, Y, Gou, W, Wu, P, Lai, Y, Liang, X, Zhang, K, et al.. Landscape of the gut mycobiome dynamics during pregnancy and its relationship with host metabolism and pregnancy health. Gut 2024;73:1302–12. https://doi.org/10.1136/gutjnl-2024-332260.Search in Google Scholar PubMed PubMed Central
42. Hu, H, Ma, J, Peng, Y, Feng, R, Luo, C, Zhang, M, et al.. Thrombospondin-1 regulates trophoblast necroptosis via NEDD4-mediated ubiquitination of TAK1 in preeclampsia. Adv Sci (Weinh) 2024;11:e2309002. https://doi.org/10.1002/advs.202309002.Search in Google Scholar PubMed PubMed Central
43. Rahnavard, A, Chatterjee, R, Wen, H, Gaylord, C, Mugusi, S, Klatt, KC, et al.. Molecular epidemiology of pregnancy using omics data: advances, success stories, and challenges. J Transl Med 2024;22:106. https://doi.org/10.1186/s12967-024-04876-7.Search in Google Scholar PubMed PubMed Central
44. Haniffa, M, Maartens, A, Winheim, E, Jardine, L. Decoding the human prenatal immune system with single-cell multi-omics. Nat Rev Immunol 2025;25:285–97. https://doi.org/10.1038/s41577-024-01099-1.Search in Google Scholar PubMed
45. Doratt, BM, True, HE, Sureshchandra, S, Qiao, Q, Rincon, M, Marshall, NE, et al.. The immune landscape of fetal chorionic villous tissue in term placenta. Front Immunol 2025;15:1506305. https://doi.org/10.3389/fimmu.2024.1506305.Search in Google Scholar PubMed PubMed Central
46. Lea, G, Doria-Borrell, P, Ferrero-Micó, A, Varma, A, Simon, C, Anderson, H, et al.. Ectopic expression of DNMT3L in human trophoblast stem cells restores features of the placental methylome. Cell Stem Cell 2025;32:276–92.e9. https://doi.org/10.1016/j.stem.2024.12.007.Search in Google Scholar PubMed
47. Wu, SPS, Quiroz, E, Wang, T, Montague Redecke, SG, Xu, X, Lin, L, et al.. Assessment of the histone mark-based epigenomic landscape in human myometrium at term pregnancy. eLife 2025;13:RP95897. https://doi.org/10.1101/2024.02.19.581035.Search in Google Scholar PubMed PubMed Central
48. Friedman, C, Niemiec, S, Dabelea, D, Kechris, K, Yang, IV, Adgate, JL, et al.. Prenatal black carbon exposure and DNA methylation in umbilical cord blood. Int J Hyg Environ Health 2025;263:114464. https://doi.org/10.1016/j.ijheh.2024.114464.Search in Google Scholar PubMed PubMed Central
49. Barcelona, V, Ray, M, Zhao, Y, Samari, G, Wu, H, Reho, P, et al.. Epigenomic pathways from racism to preterm birth: secondary analysis of the Nulliparous Pregnancy Outcomes Study cohort in the USA. BMJ Open 2025;15:e091801. https://doi.org/10.1136/bmjopen-2024-091801.Search in Google Scholar PubMed PubMed Central
50. van Vliet, MM, Schoenmakers, S, Boers, RG, van der Meeren, LE, Gribnau, J, Steegers-Theunissen, RPM. Methylome profiling of cell-free DNA during the early life course in (un)complicated pregnancies using MeD-seq: protocol for a cohort study. PLoS One 2025;20:e0310019. https://doi.org/10.1371/journal.pone.0310019.Search in Google Scholar PubMed PubMed Central
51. Li, L, Baek, KH. Exploring potential biomarkers in recurrent pregnancy loss: a literature review of omics studies to molecular mechanisms. Int J Mol Sci 2025;26:2263. https://doi.org/10.3390/ijms26052263.Search in Google Scholar PubMed PubMed Central
52. Mumcu, A, Sarıdoğan, E, Düz, SA, Tuncay, G, Erdoğan, A, Karaer, K, et al.. Multi-omics analysis of placental metabolomics and transcriptomics datasets reveals comprehensive insights into the pathophysiology of preeclampsia. J Pharm Biomed Anal 2025;256:116701. https://doi.org/10.1016/j.jpba.2025.116701.Search in Google Scholar PubMed
53. Puvvula, J, Song, LC, Zalewska, KJ, Alexander, A, Manz, KE, Braun, JM, et al.. Global metabolomic alterations associated with endocrine-disrupting chemicals among pregnant individuals and newborns. Metabolomics 2025;21:20. https://doi.org/10.1007/s11306-024-02219-7.Search in Google Scholar PubMed PubMed Central
54. Shi, Y, Li, K, Ding, R, Li, X, Cheng, Z, Liu, J, et al.. Untargeted metabolomics and machine learning unveil the exposome and metabolism linked with the risk of early pregnancy loss. J Hazard Mater 2025;488:137362. https://doi.org/10.1016/j.jhazmat.2025.137362.Search in Google Scholar PubMed
55. Okuda, K, Nagano, N, Nakazaki, K, Matsuda, K, Tokunaga, W, Fuwa, K, et al.. Metabolomic profiles of preterm small-for-gestational age infants. Pediatr Neonatol 2025;66:50–4. https://doi.org/10.1016/j.pedneo.2023.11.012.Search in Google Scholar PubMed
56. Chen, W, Qiu, C, Hao, J, Liao, J, Lurmann, F, Pavlovic, N, et al.. Maternal metabolomics linking prenatal exposure to fine particulate matter and birth weight: a cross-sectional analysis of the MADRES cohort. Environ Health 2025;24:14. https://doi.org/10.1186/s12940-025-01162-x.Search in Google Scholar PubMed PubMed Central
57. Meng, X, Shi, M, Qian, J, Luo, Y, Cui, L, Li, D, et al.. Tacrolimus (FK506) promotes placentation and maternal-fetal tolerance through modulating FASN-CEACAM1 pathway. Front Immunol 2025;16:1522346. https://doi.org/10.3389/fimmu.2025.1522346.Search in Google Scholar PubMed PubMed Central
58. Nonn, O, Debnath, O, Valdes, DS, Sallinger, K, Secener, AK, Fischer, C, et al.. Senescent syncytiotrophoblast secretion during early onset preeclampsia. Hypertension 2025;82:787–99. https://doi.org/10.1161/HYPERTENSIONAHA.124.23362.Search in Google Scholar PubMed PubMed Central
59. Li, S, Zhang, Y, Yuan, R, Zhu, S, Bai, J, Miao, Y, et al.. ARHGAP26 deficiency drives the oocyte aneuploidy and early embryonic development failure. Cell Death Differ 2025;32:291–305. https://doi.org/10.1038/s41418-024-01384-5.Search in Google Scholar PubMed PubMed Central
60. Cyr-Depauw, C, Mižik, I, Cook, DP, Lesage, F, Vadivel, A, Messaoudi, I, et al.. Single-cell RNA sequencing to guide autologous preterm cord mesenchymal stromal cell therapy. Am J Respir Crit Care Med 2025;211:391–406. https://doi.org/10.1164/rccm.202403-0569OC.Search in Google Scholar PubMed
61. Luo, Y, An, C, Zhong, K, Zhou, P, Li, D, Liu, H, et al.. Exploring the impacts of senescence on implantation and early embryonic development using totipotent cell-derived blastoids. J Adv Res 2025;68:115–29. https://doi.org/10.1016/j.jare.2024.02.011.Search in Google Scholar PubMed PubMed Central
62. Hu, H, Peng, Y, Wang, CC, Chen, J, Yu, X, Chen, X, et al.. Neutrophil extracellular traps induce trophoblasts pyroptosis via enhancing NLRP3 lactylation in SLE pregnancies. J Autoimmun 2025;153:103411. https://doi.org/10.1016/j.jaut.2025.103411.Search in Google Scholar PubMed
63. d’Errico, A, Popovic, M, Pizzi, C, Moirano, G, Moccia, C, Richiardi, L, et al.. Maternal occupational exposures during early stages of pregnancy and adverse birth outcomes in the NINFEA birth cohort. PLoS One 2025;20:e0313085. https://doi.org/10.1371/journal.pone.0313085.Search in Google Scholar PubMed PubMed Central
64. Li, X, Ran, M, Wang, M, Liu, A, Qiao, B, Han, B, et al.. PM2.5 constituent exposures and maternal circulatory homocysteine in early pregnancy. Environ Health 2025;24:7. https://doi.org/10.1186/s12940-025-01160-z.Search in Google Scholar PubMed PubMed Central
65. Lakhoo, DP, Brink, N, Radebe, L, Craig, MH, Pham, MD, Haghighi, MM, et al.. A systematic review and meta-analysis of heat exposure impacts on maternal, fetal and neonatal health. Nat Med 2025;31:684–94. https://doi.org/10.1038/s41591-024-03395-8.Search in Google Scholar PubMed PubMed Central
66. Xie, Y, Xiao, H, Zheng, D, Mahai, G, Li, Y, Xia, W, et al.. Associations of prenatal metal exposure with child neurodevelopment and mediation by perturbation of metabolic pathways. Nat Commun 2025;16:2089. https://doi.org/10.1038/s41467-025-57253-3.Search in Google Scholar PubMed PubMed Central
67. Nakamura, K. Immunotoxicological disruption of pregnancy as a new research area in immunotoxicology. J Immunot 2025;22:2475772. https://doi.org/10.1080/1547691X.2025.2475772.Search in Google Scholar PubMed
68. Andrade, C. Autism spectrum disorder, 1: genetic and environmental risk factors. J Clin Psychiatry 2025;86:25f15878. https://doi.org/10.4088/JCP.25f15878.Search in Google Scholar PubMed
69. Chen, Z, Zheng, M, Wan, T, Li, J, Yuan, X, Qin, L, et al.. Gestational exposure to nanoplastics disrupts fetal development by promoting the placental aging via ferroptosis of syncytiotrophoblast. Environ Int 2025;197:109361. https://doi.org/10.1016/j.envint.2025.109361.Search in Google Scholar PubMed
70. Panneerselvam, D, Murugesan, A, Raveendran, SK, Kumar, JS, Venkataraman, P. Examining the hidden dangers: understanding how microplastics affect pregnancy. Eur J Obstet Gynecol Reprod Biol 2025;304:53–62. https://doi.org/10.1016/j.ejogrb.2024.11.024.Search in Google Scholar PubMed
71. Peeples, ES, Molloy, EJ, Bearer, CF. Novel biomarkers of fetal and neonatal environmental exposure, effect and susceptibility. Pediatr Res 2025;98:813–8. https://doi.org/10.1038/s41390-025-03816-5.Search in Google Scholar PubMed PubMed Central
72. Veras, MM, Saldiva, PHN. Impact of air pollution and climate change on maternal, fetal and postnatal health. J Pediatr (Rio J). 2025;101:S48–55. https://doi.org/10.1016/j.jped.2024.10.006.Search in Google Scholar PubMed PubMed Central
73. Kobayashi, S, Saijo, Y, Itoh, M, Tamura, N, Tojo, M, Iwata, H, et al.. Effects of the maternal work environment on psychological distress during pregnancy: the Japan Environment and Children’s Study. J Occup Environ Med 2025;67:89–99. https://doi.org/10.1097/JOM.0000000000003276.Search in Google Scholar PubMed
74 Cai, R, Yang, Q, Liao, Y, Qin, L, Han, J, Gao, R. Immune treatment strategies in unexplained recurrent pregnancy loss. Am J Reprod Immunol. 2025;93:e70060. https://doi.org/10.1111/aji.70060.Search in Google Scholar PubMed
75. Monticciolo, I, Guarano, A, Inversetti, A, Barbaro, G, Di Simone, N. Unexplained recurrent pregnancy loss: clinical application of immunophenotyping. Am J Reprod Immunol 2024;92:e13939.10.1111/aji.13939Search in Google Scholar PubMed
76. Jiang, L, Zhang, L, Xia, J, Cheng, L, Chen, G, Wang, J, et al.. Probiotics supplementation during pregnancy or infancy on multiple food allergies and gut microbiota: a systematic review and meta-analysis. Nutr Rev 2025;83:e25–41. https://doi.org/10.1093/nutrit/nuae024.Search in Google Scholar PubMed PubMed Central
77. Matsumura, K, Hamazaki, K, Tsuchida, A, Inadera, H; Japan Environment and Children’s Study (JECS) Group, Yamazaki, S, et al.. Prospective association of air purifier use during pregnancy with the neurodevelopment of toddlers in the Japan Environment and Children’s Study. Sci Rep 2021;11:19454. https://doi.org/10.1038/s41598-021-98482-y.Search in Google Scholar PubMed PubMed Central
78. Orchanian, SB, Hsiao, EY. The microbiome as a modulator of neurological health across the maternal-offspring interface. J Clin Investig 2025;135:e184314. https://doi.org/10.1172/JCI184314.Search in Google Scholar PubMed PubMed Central
79. Xie, Y, Chen, Q, Shan, D, Pan, X, Hu, Y. Unraveling the role of the gut microbiome in pregnancy disorders: insights and implications. Front Cell Infect Microbiol 2025;15:1521754. https://doi.org/10.3389/fcimb.2025.1521754.Search in Google Scholar PubMed PubMed Central
80. Enache, RM, Roşu, OA, Profir, M, Pavelescu, LA, Creţoiu, SM, Gaspar, BS. Correlations between gut microbiota composition, medical nutrition therapy, and insulin resistance in pregnancy: a narrative review. Int J Mol Sci 2025;26:1372. https://doi.org/10.3390/ijms26031372.Search in Google Scholar PubMed PubMed Central
81. Nardi, E, Seidita, I, Abati, I, Donati, C, Bernacchioni, C, Castiglione, F, et al.. The placenta in fetal death: molecular evidence of dysregulation of inflammatory, proliferative, and fetal protective pathways. Am J Obstet Gynecol 2025;232:328.e1–328.e9. https://doi.org/10.1016/j.ajog.2024.06.011.Search in Google Scholar PubMed
82. Llorca, T, Ruiz-Magaña, MJ, Abadía, AC, Ruiz-Ruiz, C, Olivares, EG. Decidual stromal cells: fibroblasts specialized in immunoregulation during pregnancy. Trends Immunol 2025;46:138–52. https://doi.org/10.1016/j.it.2024.12.007.Search in Google Scholar PubMed
83. Li, B, Xiong, Y, Guo, D, Deng, G, Wu, H. The gut-reproductive axis: bridging microbiota balances to reproductive health and fetal development. Int Immunopharmacol 2025;144:113627. https://doi.org/10.1016/j.intimp.2024.113627.Search in Google Scholar PubMed
84. Dang, H, Feng, P, Zhang, S, Peng, L, Xing, S, Li, Y, et al.. Maternal gut microbiota influence stem cell function in offspring. Cell Stem Cell 2025;32:246–62.e8. https://doi.org/10.1016/j.stem.2024.10.003.Search in Google Scholar PubMed
85. Gu, B, Ferreira, LMR, Herrera, S, Brown, L, Lieberman, J, Sherwood, RI, et al.. The TEA domain transcription factors TEAD1 and TEAD3 and WNT signaling determine HLA-G expression in human extravillous trophoblasts. Proc Natl Acad Sci U S A. 2025;122:e2425339122. https://doi.org/10.1073/pnas.2425339122.Search in Google Scholar PubMed PubMed Central
86. Cai, S, Xue, B, Li, S, Wang, X, Zeng, X, Zhu, Z, et al.. Methionine regulates maternal-fetal immune tolerance and endometrial receptivity by enhancing embryonic IL-5 secretion. Cell Rep 2025;44:115291. https://doi.org/10.1016/j.celrep.2025.115291.Search in Google Scholar PubMed
87. Acharya, B, Behera, A, Moharana, S, Prajapati, BG, Behera, S. Nanoparticle-mediated embryotoxicity: mechanisms of chemical toxicity and implications for biological development. Chem Res Toxicol 2025;38:521–41. https://doi.org/10.1021/acs.chemrestox.4c00472.Search in Google Scholar PubMed
88. He, L, Zheng, S, Zhan, F, Lin, N. The role of necroptosis in pathological pregnancies: mechanisms and therapeutic opportunities. J Reprod Immunol 2025;169:104460. https://doi.org/10.1016/j.jri.2025.104460.Search in Google Scholar PubMed
89. Nong, Y, Zhai, Q, Liu, W, Wei, J, Wang, Z, Lv, X, et al.. AQP3 influences unexplained recurrent abortion by regulating trophoblast cell migration and invasion via the METTL14/IGF2BP1/AQP3/PI3K/AKT pathway. J Cell Mol Med 2025;29:e70325. https://doi.org/10.1111/jcmm.70325.Search in Google Scholar PubMed PubMed Central
90. Shukla, AK, Ahamad, S, Kukshal, P. Computational insights into maternal environmental pollutants and folate pathway regulation. Reprod Toxicol 2025;132:108825. https://doi.org/10.1016/j.reprotox.2024.108825.Search in Google Scholar PubMed
91. Singh, S, Goel, I, Quadri, JA, Minocha, R, Kashyap, N, Rana, A, et al.. Environmental pollutant SO2 exposure affects trophoblast function involving an ER stress pathway. J Physiol 2025;603:1263–79. https://doi.org/10.1113/JP287409.Search in Google Scholar PubMed
92. Moss, CG, Dilworth, MR, Harris, LK, Freeman, S, Heazell, AEP. Understanding a potential role for the NLRP3 inflammasome in placenta-mediated pregnancy complications. Am J Reprod Immunol 2025;93:e70077. https://doi.org/10.1111/aji.70077.Search in Google Scholar PubMed PubMed Central
93. Muse, ME, Wang, Y, Gilbert-Diamond, D, Armstrong, DA, Hoen, AG, Romano, ME, et al.. Maternal diet quality and circulating extracellular vesicle and particle miRNA during pregnancy. Eur J Nutr 2025;64:75. https://doi.org/10.1007/s00394-025-03589-x.Search in Google Scholar PubMed PubMed Central
94. Zhang, M, Li, K, Huang, X, Zhou, H, Tan, J, Guo, Z, et al.. Gene expression profiles based on maternal plasma cfDNA nucleosome footprints indicate fetal development and maternal immunity changes during pregnancy progress. Placenta 2025;159:84–92. https://doi.org/10.1016/j.placenta.2024.12.005.Search in Google Scholar PubMed
95. Wang, T, Sun, L, Li, M, Zhang, Y, Huang, L. Transcriptomics reveals preterm birth risk: identification and validation of key genes in monocytes. BMC Pregnancy Childbirth 2025;25:174. https://doi.org/10.1186/s12884-025-07293-w.Search in Google Scholar PubMed PubMed Central
96. Kong, L, Yang, H, Yang, J, Jiang, L, Xu, B, Yang, T, et al.. Role of calcium overload-mediated disruption of mitochondrial dynamics in offspring neurotoxicity due to methylmercury exposure during pregnancy and lactation. Ecotoxicol Environ Saf 2025;291:117835. https://doi.org/10.1016/j.ecoenv.2025.117835.Search in Google Scholar PubMed
97. Neumann, A, Sammallahti, S, Cosin-Tomas, M, Reese, SE, Suderman, M, Alemany, S, et al.. Epigenetic timing effects on child developmental outcomes: a longitudinal meta-regression of findings from the Pregnancy and Childhood Epigenetics Consortium. Genome Med 2025;17:39. https://doi.org/10.1186/s13073-025-01451-7.Search in Google Scholar PubMed PubMed Central
98. Sierla, JR, Pagerols Raluy, L, Trochimiuk, M, Trah, J, Petrosyan, M, Velasquez, LN, et al.. Disparity between functional and structural recovery of placental mitochondria after exposure to hypoxia. Int J Mol Sci 2025;26:2956. https://doi.org/10.3390/ijms26072956.Search in Google Scholar PubMed PubMed Central
99. Adibi, JJ, Zhao, Y, Koistinen, H, Mitchell, RT, Barrett, ES, Miller, R, et al.. Molecular pathways in placental-fetal development and disruption. Mol Cell Endocrinol 2024;581:112075. https://doi.org/10.1016/j.mce.2023.112075.Search in Google Scholar PubMed PubMed Central
100. Liu, L, Tang, L, Chen, S, Zheng, L, Ma, X. Decoding the molecular pathways governing trophoblast migration and placental development: a literature review. Front Endocrinol (Lausanne) 2024;15:1486608. https://doi.org/10.3389/fendo.2024.1486608.Search in Google Scholar PubMed PubMed Central
101. Scher, MS. The neural exposome influences the preterm fetal-to-neonatal connectome. Pediatr Res 2024;95:9–11. https://doi.org/10.1038/s41390-023-02804-x.Search in Google Scholar PubMed
© 2025 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.


