Abstract
The three-dimensional (3D) reconstruction of medical images usually requires hundreds of two-dimensional (2D) scan images. Segmentation, an obligatory part in reconstruction, needs to be performed for all the slices consuming enormous storage space and time. To reduce storage space and time, this paper proposes a three-stage procedure, namely, slice selection, segmentation and interpolation. The methodology will have the potential to 3D reconstruct the human head from minimum selected slices. The first stage of slice selection is based on structural similarity measurement, discarding the most similar slices with none or minimal impact on details. The second stage of segmentation of the selected slices is performed using our proposed phase-field segmentation method. Validation of our segmentation results is done via comparison with other deformable models, and results show that the proposed method provides fast and accurate segmentation. The third stage of interpolation is based on modified curvature registration-based interpolation, and it is applied to re-create the discarded slices. This method is compared to both standard linear interpolation and registration-based interpolation in 100 tomographic data sets. Results show that the modified curvature registration-based interpolation reconstructs missing slices with 96% accuracy and shows an improvement in sensitivity (95.802%) on par with specificity (95.901%).
1 Introduction
Since the advent of non-invasive imaging technology such as computed tomography (CT), visualization of cross-sectional images or internal anatomical structures has been fast, and accurate analysis of bones, muscles, fats and organs in order to treat injuries/abnormalities such as skull fractures, space-occupying lesions, intracranial hemorrhage, midline shift, hydrocephalus, ischemia, etc., has become easier for medical practitioners. These two-dimensional (2D) CT slices comprise large volumes of complex clinical database. A high-resolution CT scanner has a narrow slice thickness of usually 1–2 mm, and spacing between the slices can vary between 1 and 15 mm depending upon the organ being imaged. Visualizing and analyzing these 2D slices individually for identification of the diseased portion are a tedious and time-consuming task also prone to subjective biases.
Computer-assisted, three-dimensional (3D) geometric models generated using volumetric reconstruction have, no doubt, been aiding the surgeons improve the diagnosis. However, the huge and constant accumulation in the volume and variety of CT slices has created a severe threat in terms of storage required and is a problem that needs to be addressed.
The 3D volumetric human head reconstruction from 2D CT slices requires segmenting all the hundreds of slices, which consumes time and storage. Reduction of the number of slices to be segmented is possible if the adjacent slices are closely spaced and exhibit a good amount of similarity. As the images chosen are medical images, there exists a risk of omitting details/organs partially or completely if the removal of slices is not done carefully. Considering all these aspects and the requirement of reducing the storage space, this paper is developed and comprises three stages, namely, (1) slice selection, (2) segmentation of selected slices and (3) slice reconstruction. The term “slice reconstruction” in this paper refers to the process of re-creating the discarded slices using interpolation and should not be confused with 3D image reconstruction. The first stage is applying an objective measure called the structural similarity measure (SSIM), which is a measure of perceived structural information change [43] to calculate the similarity between adjacent head CT slices. This image quality assessment includes visual perception along with luminance and contrast masking unlike the traditional methods like mean square error (MSE) and peak signal-to-noise ratio (PSNR), which estimate only absolute errors [12, 40, 41, 42]. The mean SSIM provides a higher value of 1 for totally similar slices and zero otherwise. Consequently, only those slices that do not exhibit a higher similarity value with the adjacent slice need to be selected for segmentation discarding the most similar ones. Segmentation of the selected slices forms the second stage.
In medical image processing, segmentation of tissues/organs or extracting the region of interest either automatically or semi-automatically are, by far, the most extensively researched topics. Some of the techniques for segmentation in the literature include the region growing, clustering, thresholding, deformable models, neural networks, fuzzy-based approaches, etc. Here, we briefly review related works on deformable model [15, 29] based segmentation. A detailed review on various segmentation techniques can be found in Refs. [32] and [34]. Two important classifications of deformable models exist, namely, the parametric model, which represents curves and surfaces explicitly, and the geometric model that represents curves and surfaces implicitly as a level set function considering the topological changes, too. The active contour model (ACM) is an iterative energy minimization parametric model. This snake model [17] is a set of connected points guided by the user-defined external constraint forces, internal forces and image forces to pull it toward the edges by minimizing the energy function. The problem of poor concavity and sensitivity to initialization with ACM is well addressed in Refs. [7, 35, 44] by introducing a new class of external forces called gradient vector flow (GVF), which detects boundary concavities, shows a large capture range, and insensitivity to initialization. The geodesic active contour model (GACM) comes under the geometric model [5, 31], which uses a gradient flow as a boundary stopping function, and the high variation in gradient attracts the curve to the boundary. A model based on regions using Mumford Shah functional and level sets known as active contours without edges or the Chan Vese (CV) model is proposed in Ref. [6]. It can detect edges without the need for gradient. Because of the frequent occurrence of heterogeneous objects in medical imagery, another region-based segmentation method known as localizing region-based active contours (LRAC) is explained in Ref. [18]. It utilizes the local information for the contour to evolve by considering the local image statistics rather than global ones. The distance regularized level set evolution (DRLSE) is a variation provided to the level set. It is derived with a distance-regularized term that helps in minimizing the energy functional and an external energy term that controls the motion of zero level set to the desired boundary [25], thereby, avoiding re-initialization.
Benes et al. [3] are the first to have suggested an alternative to the level set motion by mean curvature flow based on phase field approach. This geometrical image segmentation is based on the numerical solution of the Allen-Cahn (AC) equation. Another robust and accurate geometric active contour model (ACM) for segmentation of artificial as well as medical images based on modified AC equation and phase field approximation is discussed in Ref. [22]. The initial guess for this segmentation is given in the form of a characteristic function covering the object of interest and the edge stopping function used stops the evolution when the contour reaches the edge. Refs. [23, 24, 26] discuss a stable hybrid numerical method for bimodal image segmentation, multiphase image segmentation and a stable numerical method for solving the Allen-Cahn equation. An efficient detection of edges/boundaries in human bone slices is mentioned in Ref. [27] using the modified AC equation and phase-field function. This iterative method is named as the iterative phase-field segmentation (IPFS). An improvisation to this method in terms of reducing the number of iterations is proposed in this paper, and the proposed method is, henceforth, referred to as the phase-field segmentation (PFS).
This PFS segmentation is applied only to the slices that have been selected based on mean SSIM. In the third stage, the discarded slices in the first stage are re-created using interpolation for the purpose of 3D reconstruction. Interpolation is the process of defining a function that takes on specified values at specified points or guessing the intensity values at missing locations. Finding a suitable interpolation technique specific to the image/application is essential as image quality highly depends on it. Spatial interpolation is classified into object-based and intensity-based interpolation. Linear and cubic interpolation calculates the weighted average of the intensity values of input images and is computationally simple. Object-based interpolation provides more accurate interpolation by extracting additional information contained in the image slices. There are a number of interpolation techniques proposed in the literature. An earlier interpolation method highly influenced by the shape of the object is the shape-based interpolation [36]. Here, the binary image obtained after segmentation is subjected to a distance transform, which results in the creation of gray-level data. This gray value (positive for points inside the object and negative outside) at each point represents the shortest distance from the boundary, and this gray image is interpolated. The interpolated image with a positive set of points forms the interpolated object having been thresholded at zero. This shape-based interpolation method has been extended and generalized in Ref. [13] for gray-level input data sets taking into account the shape of the intensity distribution profile in the multidimensional space. However, it cannot effectively deal with objects with holes or large offsets. To overcome this drawback, feature-guided shape-based image interpolation [19] has been developed. An objective comparison of 3D image interpolation methods is given in Ref. [14]. Other methods for interpolation include the morphology-based method by Lee and Wang [20], morphing-based method for medical images [1], registration-based interpolation [11, 33], etc.
Image registration is aligning one image to another so that they share the same coordinate system. This method needs to estimate an optimal transformation model that minimizes the variance between images. Given two images, a reference R and a template T, the image registration algorithm [30] finds a spatial transformation, such that the transformation optimizes the image alignment criterion. The criterion is usually a cost function or similarity measure describing the spatial alignment of features representing the registration basis. However, the actual choice of the transformation model and similarity metric depends on the application under consideration. Maintz and Viergever [28] classified the registration algorithms based on dimensionality, type of modalities used, nature of the registration basis, geometrical transformation, user interaction and type of optimization procedure. A review on medical image registration emphasizing intrasubject registration of tomographic modalities is given in Ref. [16].
The well-known transformation models that relate the position of features in two images with 6 and 12 degrees of freedom are the rigid and affine transformation. Spline-based registration algorithms [8] use control points in the reference and target image and a spline function to define correspondences away from these points. B-splines are defined in the vicinity of each control point providing local support. B-spline-based non-rigid registration technique for 3D contrast-enhanced breast magnetic resonance imaging (MRI) using a combination of both affine transformation and free-form deformation based on B-spline is explained in Ref. [37]. Utilizing this non-rigid registration technique, Penny et al. [33] proposed a registration (REG)-based interpolation method to interpolate between neighboring gray-scale CT slices. Another novel REG-based interpolation uses bicubic B-spline function [21] for transformation to match the image features. By adopting a multi-resolution strategy, the features are matched from coarse to fine. A non-linear REG model [9, 10] based on curvature type smoother computes the internal forces so as to minimize the curvature of the displacement field, thereby, allowing automatic affine linear alignment, thus, avoiding a pre-REG step. Also, a fast and direct solver is developed based on real discrete cosine transformation. Ref. [2] uses this curvature-based REG method for slice interpolation of medical images by considering a linear displacement between corresponding pixels of reference and template image. The displacement field computed is applied to both the reference and template image, and the average of the two deformed images gives the missing slice. As the movement is with respect to both the images, the computational time is reduced compared to curvature REG. This method is, hereafter, called as the modified curvature registration (MCR)-based interpolation in this paper.
Li et al. [27] applied IPFS to all the slices and used linear interpolation to insert additional slices between any two consecutive slices. This results in a larger number of slices than the original. Also, no specific criterion is mentioned regarding the removal of similar slices, which is done after segmentation and insertion. Our paper differs from Li’s paper in the following ways.
Removal of a similar slice is done based on the mean SSIM. The proposed PFS is applied only to the selected slices rather than all the slices from the data set, thus, reducing storage and time.
A comparative analysis of linear, REG-based interpolation, and MCR-based interpolation is carried out in order to re-create the reduced slices in between the PFS slices without inserting any additional slices. This is to find an appropriate and accurate interpolation technique without loss of information and quality for the purpose of a smooth 3D volumetric reconstruction of the human head.
Linear interpolation was chosen as a basic reference interpolation method; REG-based interpolation was chosen as it has been shown to have the best performance in comparison with the shape-based method [33]. The remainder of this paper is organized as follows: In Section 2, we describe the methodology used for reducing slices, segmentation and interpolation. Section 3 includes the results and discussions based on a comparative analysis of interpolation methods. Finally, our conclusion is given in Section 4.
2 Methodology
The graphical abstract of the proposed system is given in Figure 1, and the equations and methods adopted for slice removal, segmentation and interpolation are discussed in this section.

Graphical Abstract of the Proposed System.
2.1 Slice removal based on SSIM
Consider x={xi|i=1, 2, …, N} and y={yi|i=1, 2, …, N} as two discrete non-negative image signals with xi and yi representing the intensities of corresponding pixels in an image with window size N. The SSIM index [43] is,
where
The mean SSIM used to calculate the image quality of the entire image is given by,
where X and Y are the two images whose similarities are to be computed. xj, yj are the image contents at the jth local window, M is the number of samples in the quality map.
The extent of similarity between adjacent head CT slices in DICOM format is calculated using MSSIM. The process of slice selection from a set of hundreds of original head slices is done in two steps. First, a threshold is set for each five slices by averaging the similarity measures of these five slices. The slice that has a mean SSIM greater than the calculated threshold is discarded. Second, a further reduction in the ratio 2:1 is made just to show the effectiveness of the interpolation used. In order to rule out the possibility of organs disappearing in the selected slices, it is ensured that the number of slices discarded between two selected slices is limited to three. A uniform threshold is not practicable for medical images, as there is a possibility of sequential removal of slices, which may, in turn, lead to the missing details.
2.2 Iterative Phase-Field Segmentation Using Modified Allen-Cahn Equation
The phase-field model is a mathematical model for solving interfacial problems [3, 22]. The phase field or order parameter (ϕ) is used to separate one phase from the other. The edge-stopping function is calculated from the gradient image after normalization and Gaussian smoothing. The edge-stopping function plays a very vital role because this stops the evolution when the contour reaches the edge. The geometric ACM based on mean curvature motion using modified AC equation in the 2D domain Ω=(0, Lx)×(0, Ly) with x=(x, y) as its coordinate is given by:
where φ(x, t) is the phase-field function, f0(x) is the normalized value of the given image f(x), g(f0(x)) is the edge-stopping function, F(ϕ)=0.25(ϕ2−1)2 is the double well potential function, Δφ(x, t) is the Laplacian operator, ϵ>0 is a constant called phase transition width (usually set to a value ≪1), and β is a parameter chosen as 50,000.
Consider the 2D space, Ω=(a, b)×(c, d) for discretization of equation (3). The uniform mesh size is given by Δx=(b−a)/Nx and Δy=(d−c)/Ny, where Nx, Ny are positive even integers. For simplicity, Δx=Δy=h. Let
where (Gσ∗f0)x,ij =[(Gσ∗f0)i+1,j −(Gσ∗f0)i−1,j ]/2h and (Gσ∗f0)y,ij =[(Gσ∗f0)i,j+1−(Gσ∗f0)i,j−1]/2h.
Gσ is the Gaussian function (σ=1.5). F(ϕ) takes 0.25 for ϕ=0 and zero for −1 and +1 values of ϕ. Zero Neumann boundary condition is used. Equation (3) is discretized as:
Δd is a discrete five point Laplacian operator. The multigrid method [4, 39] is a fast solver for solving discretized partial differential equations, and hence, Equation (5) is solved using a multigrid method with an initial condition φn. The equation
The initial value of phase-field function is assigned to be 1 and −1 for values inside and outside the square contour. The square contour can be placed close to the object of interest by any initialization technique to speed up the evolution. Initially, F(φ) becomes zero in Equation (5), and after further iterations, φ takes a zero value on the boundary, −1 outside, and +1 inside the boundary. Equations (5) and (7) help in evolving the contour beyond non-convex and disconnected regions.
2.3 Proposed Phase-Field Segmentation
We hereby propose a PFS method for gray images by speeding up the simulation of IPFS (given in Section 2.2) by replacing the Laplacian term
2.4 Re-creation of Omitted Slices Using Linear Interpolation
Re-creation is the reconstruction of omitted slices. Linear interpolation is applied to the selected segmented slices to reconstruct the missing slices. Consider Ns to be the number of slices, and φ(x, y, zi) the segmented slice data with i=1, 2….Ns−1. Linear interpolation [27] is achieved using:
where 0≤θ≤1. Depending on the value of θ, the number of intermediate slices can be obtained. If θ=0.5, one slice will be created between two consecutive slices, if θ=0.33, two slices, if θ=0.25, three slices, and so on. Linear interpolation, even though computationally simple, creates blurred edges as it gives the weighted average of input slices.
2.5 Re-creation of Omitted Slices Using Registration-Based Interpolation
The REG-based interpolation algorithm uses non-rigid REG based on B-splines to preserve the spatial correspondence between adjacent slices. It deforms an image by manipulating a regular grid of control points based on a set of B-splines that are locally controlled. The similarity measure used to measure the degree of alignment between two images is the sum of the squared difference as the images are imaged using the same modality. REG is achieved by minimizing the cost function associated with the smoothness of transformation and image similarity. Using the non-rigid transformation and position of image plane created between two 2D slices, the interpolation directions between the two slices are defined. Optimization is carried using gradient descent algorithm. Generally, REG-based interpolation methods provide satisfactory results provided the organs in the slices taken as reference and template should not disappear, or the consecutive slices contain similar anatomical features. Additionally, the REG method should be capable of finding a suitable transformation map to match similar features. Violation of these might lead to false correspondence maps. The REG-based interpolation algorithm applied in this paper for comparison has been extensively reviewed in literature, and we refer the reader to Refs. [33] and [37] for more details.
2.6 Re-creation of Omitted Slices Using MCR-Based Interpolation
The energy functional E for MCR-based interpolation [2] is:
Let R1 and R2 be the two input images in the domain, Ω=(0, 1)×(0, 1), x be the grid of image values, S be the smoothness term, and u be the displacement value associated with each grid point that is to be determined. The ratio r=d1/(d1+d2), where d1 and d2 are the distance from R to R1 and R to R2, where R is an arbitrary slice between R1 and R2. By considering r=0.5, the output slice is assumed to be in the middle of slices R1 and R2. For simplicity, the coefficient of u is assumed to be equal to 1 in Equation (10). However, this is not the case if more than one slice has to be re-created. α is a parameter to balance the smoothness of displacement and similarity of images. The sum of the squared difference is the distance measure D used, which is given by:
where Δ is the curvature operator. The use of curvature as the smoothness term eliminates the need for an affine linear mapping or the pre-REG step. To minimize E(u) in Equation (9), the derivative of E(u) is equated to zero, and a partial differential equation given below is obtained.
where Acurv[u]=Δ2u and
To solve the PDE, a time-stepping iteration method with u0=0 and k≥0 is considered as
The discretized version of Equation (14) is
which is obtained using a finite difference approximation. l=1, 2 is the dimension index, τ is the time step, and In is the identity matrix. The solution to Equation (15) is efficiently realized [9, 10] using the discrete cosine transform (DCT). A set of coefficients for an image of size m×n with j1=1, …m, j2=1, …n can be computed using,
Defining
The intermediate slice R is obtained by averaging both the transformed input images (R1, R2) after the optimization process of calculating the final displacement field
The displacement field calculation for curvature-based REG with joint functional E(u)=D[R, T; u)]+αS is given as T(x−u(x))=R(x). Equation (9) is a modification of this equation in such a way that in MCR, the displacement field computed is applied to both R1 and R2, thereby, reducing the computational time needed for convergence. It is essential to note that in this work, as the number of slices to be re-created between the finalized slice set varies, d1, d2 and, consequently, r also varies.
3 Results and Discussion
As mentioned earlier, the main focus of this paper is to find the best interpolation technique for head CT images. An objective evaluation of the three interpolation techniques between the selected and phase-field segmented slices have been carried out.
3.1 Data
This study employs a collection of normal human head CT images taken from specialized and well-equipped scan center utilizing Siemens SOMATOM Sensation 64-slice CT scanner (Forchheim, Germany) (cardiac imaging with rotating time of 0.33 s and isotropic spatial resolution of 0.24 mm). The original images are in DICOM format, and we have chosen a test data of 100 slices for our implementation out of which the first four slices are shown in Figure 2. The current implementation was performed in MATLAB R2009b with Intel® Pentium® CPU B940 at 2 GHz, 4 GB RAM for an image size of 256∗256.

Axial cross section of a human head CT-(A–D) corresponds to the first four slices respectively in DICOM format.
3.2 Quantitative Metrics for Evaluation
As the segments are solid and have high densities, performance metrics based on the four cardinalities are preferred, namely, accuracy (ACC), sensitivity (SEN), specificity (SPEC) along with MSE and PSNR [38] for the evaluation of both segmentation and interpolation methods. For the purpose of comparison, we perform PFS to all the 100 slices to deem them as ground truth (GT). True positive (TP) gives the number of pixels that are correctly detected as foreground, whereas false positive (FP) gives the number of pixels that are falsely detected as foreground. Similarly, true negative (TN) gives the number of pixels that are correctly detected as background, whereas false negative (FN) gives the number of pixels that are falsely detected as background.
Accuracy – It is the proportion of true results (both true positives and true negatives) in the population.
Sensitivity – It is the proportion of positives measured as such.
Specificity – It is the proportion of actual negatives, which are correctly identified as such.
MSE – It measures the cumulative squared error between the interpolated/segmented (I) and the GT image with m, n as its dimensions.
PSNR – It is a measure of reconstruction quality.
3.3 Experimental Evaluation
Based on the mean SSIM, the intermediate and finalized slice selection performed is shown in Table 1 (for 25 slices), and the number of slices selected finally is only 10 out of 25. Correspondingly, only 31 slices are selected from a set of 100 slices. As a result, segmentation needs to be applied only to these 31 slices. This method helps in freeing up a lot of storage memory. We have chosen k1=0.01, k2=0.03 arbitrarily as the SSIM index algorithm is insensitive to these variations. The local window size chosen is 8×8.
Finalized slice selection based on SSIM.
| Slice no. | Mean SSIM | Slice selection | Intermediate slice selection | Finalized slice set |
|---|---|---|---|---|
| 1 | 0.767 | 1 | 1 | 1 |
| 2 | 0.762 | – | – | – |
| 3 | 0.741 | 3 | – | – |
| 4 | 0.722 | 4 | 4 | 4 |
| 5 | 0.724 | 5 | – | – |
| 6 | 0.741 | 6 | 6 | 6 |
| 7 | 0.755 | – | – | – |
| 8 | 0.76 | 8 | – | – |
| 9 | 0.766 | 9 | 9 | 9 |
| 10 | 0.778 | – | – | – |
| 11 | 0.779 | – | – | – |
| 12 | 0.777 | 12 | – | 12 |
| 13 | 0.784 | – | – | – |
| 14 | 0.781 | 14 | 14 | 14 |
| 15 | 0.782 | – | – | – |
| 16 | 0.777 | 16 | – | 16 |
| 17 | 0.786 | – | – | – |
| 18 | 0.777 | – | – | – |
| 19 | 0.767 | 19 | 19 | 19 |
| 20 | 0.772 | 20 | – | – |
| 21 | 0.764 | 21 | 21 | 21 |
| 22 | 0.766 | – | – | – |
| 23 | 0.768 | – | – | – |
| 24 | 0.766 | 24 | – | – |
| 25 | 0.782 | 25 | 25 | 25 |
The PFS model is compared against various deformable models in literature like ACM, CV model, GACM, distance regularized level set evolution (DRLSE), localized region-based ACM (LRAC), and the state-of-the art IPFS. The computational domain is set to Ω=(0, 1)×(0, 1) with h=1/128, time step Δt=4E−3, ϵ=0.03. The steady-state solution is obtained only if the relative error
Average performance analysis of different segmentation algorithms for 25 head CT slices.
| ACC (%) | SPEC (%) | SEN (%) | MSE | PSNR | Time (s) | No. of iter./slice | |
|---|---|---|---|---|---|---|---|
| ACM | 72.535 | 72.424 | 72.892 | 0.275 | 53.955 | 32.797 | 250 |
| GACM | 72.536 | 70.98 | 92.42 | 0.275 | 53.89 | 172.321 | 1400 |
| CV | 72.54 | 70.989 | 92.357 | 0.275 | 53.891 | 8.015 | 100 |
| DRLSE | 71.468 | 69.571 | 94.26 | 0.285 | 53.723 | 21.478 | 235 |
| LRAC | 71.476 | 70.048 | 87.74 | 0.285 | 53.72 | 8.378 | 220 |
| IPFS | 88.079 | 89.284 | 74.334 | 0.119 | 57.477 | 236.13 | 108 |
| PFS | 88.079 | 89.284 | 74.334 | 0.119 | 57.477 | 0.049 | 1 |
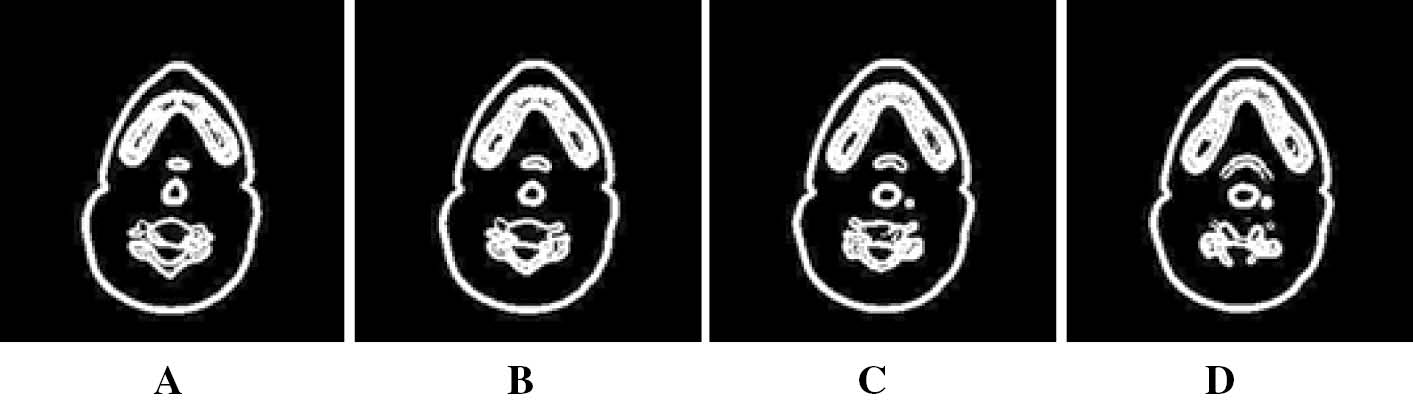
Phase Field segmented results of a human head CT. (A–D) corresponds to segmentation results of images in Figure 2.
However, when the focus is on the smooth 3D reconstruction of the human head from 2D slices, these 31 segmented slices will not suffice and it requires a greater number of slices. Interpolation helps in re-creating the discarded slices in between these 31 segmented slices. The average performance analysis for comparison of linear, REG-based interpolation and MCR interpolation for 100 slices are tabulated in Table 3. In the linear interpolation, averaging of the intensity values of the input slices is carried out which creates blurring on the edges as it does not consider the shape or structure information and, hence, is not suited for medical images.
Average performance analysis for interpolation involving 100 slices.
| ACC (%) | SPEC (%) | SEN (%) | MSE | PSNR | Time (s) | |
|---|---|---|---|---|---|---|
| Linear interpolation | 95.229 | 99.222 | 74.243 | 0.048 | 60.174 | 0.18 |
| REG interpolation | 95.369 | 98.112 | 83.397 | 0.046 | 60.434 | 162 |
| MCR interpolation | 95.948 | 95.901 | 95.802 | 0.041 | 60.918 | 30.73 |
The optimization parameter values chosen for the implementation of REG-based interpolation include a step size of 5, 2.5, 1.25 and a grid spacing of 8, 12 and 20 mm. The average time taken for REG-based interpolation is 162 s. The assumption that the objects within the input slices can only deform or move but cannot disappear should be taken into consideration, otherwise, the interpolation results become unpredictable. Moreover, this method is asymmetric as the values in the interpolated image differ depending on whether the reference image is matched to the template image or vice versa.
In the MCR-based interpolation method, the optimized displacement field for both horizontal and vertical directions is calculated, which helps in providing sharper and accurate results than linear and REG-based interpolation. We have chosen time step τ=0.05 and the regularization parameter α=100. The average computational time (30.73 s) for MCR interpolation is longer than the linear interpolation but less than the REG-based interpolation. For both REG- and MCR-based interpolation, iterations were stopped when the relative error was below 10−3. The reconstructed slices 2 and 3 between the two segmented selected slices 1 and 4 using linear, REG-based, and MCR-based interpolations are shown in Figures 4–6, respectively, for a subjective comparison. Also, it is important to note that the sensitivity (95.802%) is improved on par with specificity (95.901%) in the MCR interpolation along with the accuracy and PSNR with minimum MSE. The average performance analysis with respect to the number of slices re-created between the finalized segmented slice set using the interpolation methods discussed above is given in Table 4 for a more detailed understanding. As stated earlier, the number of slices re-created is limited to a maximum of three. As the number of slices re-created increases, there is a corresponding decrease in accuracy and PSNR with an increase in MSE. This is one of the reasons for limiting the number of slices to be re-created.

Re-creation of slices using linear interpolation. (B and C) are re-created slices by applying linear interpolation to the selected segmented slices (A and D).
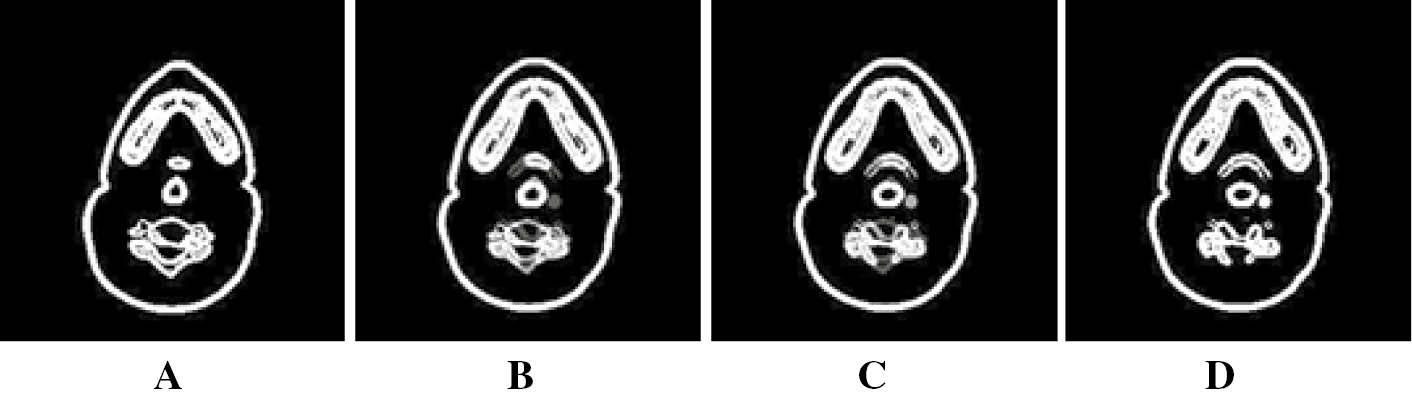
Re-creation of slices using REG based interpolation. (B and C) are re-created slices by applying REG based interpolation to the selected segmented slices (A and D).

Re-creation of slices using MCR based interpolation. (B and C) are re-created slices by applying MCR based interpolation to the selected segmented slices (A and D).
Average performance analysis based on the number of interpolated slices between two selected segmented slices.
| ACC (%) | SPEC (%) | SEN (%) | MSE | PSNR | |
|---|---|---|---|---|---|
| Single slice selection | |||||
| Linear interpolation | 95.063 | 99.209 | 72.062 | 0.05 | 61.152 |
| REG interpolation | 95.556 | 98.35 | 78.113 | 0.044 | 62.052 |
| MCR interpolation | 96.75 | 96.544 | 97.543 | 0.033 | 62.943 |
| Double slice selection | |||||
| Linear interpolation | 92.438 | 98.641 | 59.064 | 0.076 | 59.328 |
| REG interpolation | 93.996 | 97.824 | 76.176 | 0.06 | 60.876 |
| MCR interpolation | 95.313 | 94.76 | 98.101 | 0.047 | 61.397 |
| Triple slice selection | |||||
| Linear interpolation | 91.354 | 98.315 | 54.755 | 0.087 | 58.757 |
| REG interpolation | 92.647 | 96.84 | 75.555 | 0.074 | 60.01 |
| MCR interpolation | 94.021 | 93.486 | 96.591 | 0.06 | 60.338 |
Figure 7 shows the graphical plot of the performance metrics for the different interpolation methods for a set of 100 slices compared against the ground truth. The slices 1, 4, 6, 9, 12 …, being selected segmented slices and are not interpolated, show an accuracy of 100% with zero MSE and infinite PSNR, which can be visualized in the graph. The X-axis represents the 100 interpolated data set samples, and the Y-axis represents the performance metrics for evaluation. Moreover, Figure 8 depicts the performance evaluation based on the low-, medium- and high-level similarity measure. The range of low and high is below 0.6 and above 0.7, and the medium level lies in between these. This approach of measurement helps in understand better that the performance evaluation results are highly dependent on the similarity measures.

Graphs Showing Performance Analysis for Linear, Registration and MCR-based Interpolation.

Bar Graph Showing Performance Analysis Based on Three Different Levels of SSIM.
(A) Low-level SSIM. (B) Medium-level SSIM. (C) High-level SSIM. (D) SSIM-based MSE.
It may be considered that even though time reduction is achieved in segmentation, additional time is added up during interpolation. However, it must be noted that reduction in the time of segmentation in itself is justified, and as interpolation is an independent activity, time consumption may be reduced by carrying out interpolation of different slice sets in parallel. The prime focus of this work is reducing the storage requirements, and the results demonstrate very well that the requirement of storage space may be reduced to around one third.
4 Conclusion
This paper was developed to find out an accurate interpolation technique for the purpose of smooth 3D reconstruction of the human head from a set of minimally selected 2D CT slices. Minimal selection (only 31%) based on structural similarity measurement and a fast PFS algorithm for extracting the region of interest is proposed. This segmentation is applied only to these 31 selected slices, thereby, reducing time and storage required. An objective performance evaluation among the linear, REG-based and MCR-based interpolations clearly indicates that MCR-based interpolation can be used for the re-creation of omitted slices for 3D reconstruction as it significantly outperforms other methods in terms of accuracy and PSNR with reduced MSE. Our future work lies in the 3D reconstruction of human head.
Bibliography
[1] H. Atoui, S. Miguet and D. Sarrut, A fast morphing based interpolation for medical images: application to conformal radiotherapy, Image Anal. Stereol.25 (2006), 95–103.10.5566/ias.v25.p95-103Search in Google Scholar
[2] A. Baghaie and Z. Yu, Curvature-based registration for slice interpolation of medical images, computational modeling of objects presented in images fundamentals, methods, and applications, pp. 69–80, Springer International Publishing, Cham, 2014.10.1007/978-3-319-09994-1_7Search in Google Scholar
[3] M. Benes, V. Chalupecky and K. Mikula, Geometrical image segmentation by the Allen-Cahn equation, Appl. Numer. Math.51 (2004), 187–205.10.1016/j.apnum.2004.05.001Search in Google Scholar
[4] W. L. Briggs, A multigrid tutorial, SIAM, Philadelphia, Pennsylvania, 1987.Search in Google Scholar
[5] V. Caselles, R. Kimmel and G. Sapiro, Geodesic active contours, Int. J. Comput Vis.22 (1997), 61–79.10.1023/A:1007979827043Search in Google Scholar
[6] T. F. Chan and L. A. Vese, Active contour without edges, IEEE Trans. Image Process.10 (2001), 266–277.10.1109/83.902291Search in Google Scholar PubMed
[7] J. Cheng and X. Sun, Medical image segmentation with improved gradient vector flow, Res. J. Appl. Sci. Eng. Technol.4 (2012), 3951–3957.Search in Google Scholar
[8] W. R. Crum, T. Hartkens and D. L. G. Hill, Non-rigid image registration: theory and practice, Br. J. Radiol.77 (2004), S140–S153.10.1259/bjr/25329214Search in Google Scholar PubMed
[9] B. Fischer and J. Modersitzki, Curvature based image registration, J. Math. Imaging Vis.18 (2003), 81–85.10.1023/A:1021897212261Search in Google Scholar
[10] B. Fischer and J. Modersitzki, A unified approach to fast image registration and a new curvature based registration technique, Linear Algebra Appl.380 (2004), 107–124.10.1016/j.laa.2003.10.021Search in Google Scholar
[11] D. H. Frakes, L. P. Dasi, K. Pekkan, H. D. Kitajima, K. Sundareswaran, A. P. Yoganathan and M. J. T. Smith, A new method for registration-based medical image interpolation, IEEE Trans. Med. Imaging27 (2008), 370–377.10.1109/TMI.2007.907324Search in Google Scholar PubMed
[12] B. Girod, What’s wrong with mean-squared error, in: A. B. Watson, ed., Digital images and human vision, pp. 207–220, The MIT Press, Cambridge, MA, 1993.Search in Google Scholar
[13] G. J. Grevera and J. K. Udupa, Shape-based interpolation of multidimensional grey-level images, IEEE Trans Med. Imaging15 (1996), 881–892.10.1109/42.544506Search in Google Scholar PubMed
[14] G. J. Grevera and J. K. Udupa, An objective comparison of 3-D image interpolation methods, IEEE Trans Med. Imaging17 (1998), 642–652.10.1109/42.730408Search in Google Scholar PubMed
[15] L. Hi, Z. Peng, B. Everding, X. Wang, C. Y. Han, K. L. Weiss and W. G. Wee, A comparative study of deformable contour methods on medical image segmentation, Image Vis. Comput.26 (2008), 141–163.10.1016/j.imavis.2007.07.010Search in Google Scholar
[16] D. L. G. Hill, P. G. Batchelor, M. Holden and D. J. Hawkes, Medical image registration, Phys. Med. Biol.46 (2001), R1–R45.10.1088/0031-9155/46/3/201Search in Google Scholar PubMed
[17] M. Kass, A. Witkin and D. Terzopoulos, Snakes: active contour models, Proc. IEEE Int. Conf. Comput. Vis.259 (1987), 261–268.10.1007/BF00133570Search in Google Scholar
[18] S. Lankton and A. Tannenbaum, Localizing region-based active contours, IEEE Trans. Image Process. 17 (2008), 2029–2039.10.1109/TIP.2008.2004611Search in Google Scholar PubMed PubMed Central
[19] T. Y. Lee and C. H. Lin, Feature-guided shape-based image interpolation, IEEE Trans. Med. Imaging21 (2002), 1479–1489.10.1109/TMI.2002.806574Search in Google Scholar PubMed
[20] T. Y. Lee and W. H. Wang, Morphology-based three dimensional interpolation, IEEE Trans Med. Imaging19 (2000), 711–21.10.1109/42.875193Search in Google Scholar PubMed
[21] J. Leng, G. Xu and Y. Zhang, Medical image interpolation based on multi-resolution registration, Comput. Math. Appl.66 (2013), 1–18.10.1016/j.camwa.2013.04.026Search in Google Scholar
[22] Y. Li and J. Kim, A fast and accurate numerical method for medical image segmentation, J. Korean Soc. Industr. Appl. Math.14 (2010), 201–210.Search in Google Scholar
[23] Y. Li and J. Kim, Multiphase image segmentation using a phase-field model, Comput. Math. Appl.62 (2011), 737–745.10.1016/j.camwa.2011.05.054Search in Google Scholar
[24] Y. Li and J. Kim, An unconditionally stable numerical method for bimodal image segmentation, Appl. Math. Comput.219 (2012), 3083–3090.10.1016/j.amc.2012.09.038Search in Google Scholar
[25] C. Li, C. Xu, C. Gui and M. D. Fox, Distance regularized level set evolution and its application to image segmentation, IEEE Trans. Image Process.19 (2010), 3243–3254.10.1109/TIP.2010.2069690Search in Google Scholar
[26] Y. Li, H. G. Lee, D. Jeong and J. Kim, An unconditionally stable hybrid numerical method for solving the Allen–Cahn equation, Comput. Math. Appl. 60 (2010), 1591–1606.10.1016/j.camwa.2010.06.041Search in Google Scholar
[27] Y. Li, J. Shin, Y. Choi and J. Kim, Three-dimensional volume reconstruction from slice data using phase field models, Comput. Vis. Image Underst.137 (2015), 115–124.10.1016/j.cviu.2015.02.001Search in Google Scholar
[28] J. B. Maintz and M. A. Viergever, A survey of medical image registration, Med. Image Anal.2 (1998), 1–36.10.1016/S1361-8415(98)80001-7Search in Google Scholar
[29] T. McInerney and D. Terzopoulos, Deformable models in medical image analysis: a survey, Med. Image Anal.1 (1996), 91–108.10.1016/S1361-8415(96)80007-7Search in Google Scholar
[30] J. Modersitzki, Numerical methods for image registration, numerical mathematics and scientific computation, Oxford University Press, New York, NY, 2004.10.1093/acprof:oso/9780198528418.001.0001Search in Google Scholar
[31] W. J. Niessen, B. M. ter Haar Romeny and M. A. Viergever, Geodesic deformable models for medical image analysis, IEEE Trans. Med. Imaging17 (1998), 634–641.10.1109/42.730407Search in Google Scholar PubMed
[32] A. Norouzi, M. Shafry, M. Rahim, A. Altameem, T. Saba, A. E. Rad, A. Rehman and M. Uddin, Medical image segmentation methods, algorithms, and applications, IETE Tech. Rev. 31 (2014), 199–213.10.1080/02564602.2014.906861Search in Google Scholar
[33] G. P. Penney, J. A. Schnabel, D. Rueckert, M. A. Viergever and W. J. Niessen, Registration-based interpolation, IEEE Trans. Med. Imaging23 (2004), 922–926.10.1109/TMI.2004.828352Search in Google Scholar PubMed
[34] D. L. Pham, C. Xu and J. L. Prince, Current methods in medical image segmentation, Annu. Rev. Biomed. Eng.2 (2000), 315–337.10.1146/annurev.bioeng.2.1.315Search in Google Scholar PubMed
[35] L. Qin, C. Zhu, Y. Zhao, H. Bai and H. Tian, Generalized gradient vector flow for snakes: new observations, analysis and improvement, IEEE Trans. Circuits Syst. Video Technol.23(5) (2013), 883–897.10.1109/TCSVT.2013.2242554Search in Google Scholar
[36] S. P. Raya and J. K. Udupa, Shape-based interpolation of multidimensional objects, IEEE Trans. Med. Imaging9 (1990), 32–42.10.1109/42.52980Search in Google Scholar PubMed
[37] D. Rueckert, L. Sonoda, C. Hayes, D. Hill, M. Leach and D. Hawkes, Non-rigid registration using free-form deformations: application to breast MR images, IEEE Trans. Med. Imaging18 (1999), 712–721.10.1109/42.796284Search in Google Scholar PubMed
[38] A. A. H. Thasneem, R. Mehaboobathunnisa, M. M. Sathik and S. Arumugam, Comparison of different segmentation algorithms for dermoscopic images, ICTACT J. Image Video Process.05 (2015), 1030–1036.10.21917/ijivp.2015.0151Search in Google Scholar
[39] U. Trottenberg, C. Oosterlee and A. Schüller, Multigrid, Academic Press, San Diego, CA, 2001.Search in Google Scholar
[40] Z. Wang, The SSIM index for image quality assessment. http://www.cns.nyu.edu/∼lcv/ssim/.Search in Google Scholar
[41] Z. Wang and A. C. Bovik, A universal image quality index, IEEE Signal Process. Lett.9 (2002), 81–84.10.1109/97.995823Search in Google Scholar
[42] Z. Wang, A. C. Bovik and L. Lu, Why is image quality assessment so difficult, Proc. IEEE Int. Conf. Acoust. Speech Signal Process.4 (2002), 3313–3316.10.1109/ICASSP.2002.5745362Search in Google Scholar
[43] Z. Wang, A. C. Bovik, H. R. Sheikh and E. P. Simoncelli, Image quality assessment: from error measurement to structural similarity, IEEE Trans. Image Process.13 (2004), 600–612.10.1109/TIP.2003.819861Search in Google Scholar PubMed
[44] C. Xu and J. Prince, Snakes, shapes, and gradient vector flow, IEEE Trans. Image Process.7 (1998), 359–369.10.1109/83.661186Search in Google Scholar PubMed
©2019 Walter de Gruyter GmbH, Berlin/Boston
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Articles in the same Issue
- Frontmatter
- Fusion Algorithm of Multi-focus Images with Weighted Ratios and Weighted Gradient Based on Wavelet Transform
- A Novel Approach to Extract Exact Liver Image Boundary from Abdominal CT Scan using Neutrosophic Set and Fast Marching Method
- A Fast Segmentation and Efficient Slice Reconstruction Technique for Head CT Images
- Fuzzy Approach to Decision Support System Design for Inventory Control and Management
- Multiple-Reservoir Scheduling Using β-Hill Climbing Algorithm
- Combining Wavelet Texture Features and Deep Neural Network for Tumor Detection and Segmentation Over MRI
- An Efficient Adaptive Filter for Fetal ECG Extraction Using Neural Network
- A Multi-Agents System for Solving Facility Layout Problem: Application to Operating Theater
- Global Research Trends of Intuitionistic Fuzzy Set: A Bibliometric Analysis
- Nurse Scheduling with Opposition-Based Parallel Harmony Search Algorithm
- Some Innovative Types of Fuzzy Ideals in AG-Groupoids
- Extracting Conceptual Relationships and Inducing Concept Lattices from Unstructured Text
- A Hybrid Cuckoo Search and Simulated Annealing Algorithm
Articles in the same Issue
- Frontmatter
- Fusion Algorithm of Multi-focus Images with Weighted Ratios and Weighted Gradient Based on Wavelet Transform
- A Novel Approach to Extract Exact Liver Image Boundary from Abdominal CT Scan using Neutrosophic Set and Fast Marching Method
- A Fast Segmentation and Efficient Slice Reconstruction Technique for Head CT Images
- Fuzzy Approach to Decision Support System Design for Inventory Control and Management
- Multiple-Reservoir Scheduling Using β-Hill Climbing Algorithm
- Combining Wavelet Texture Features and Deep Neural Network for Tumor Detection and Segmentation Over MRI
- An Efficient Adaptive Filter for Fetal ECG Extraction Using Neural Network
- A Multi-Agents System for Solving Facility Layout Problem: Application to Operating Theater
- Global Research Trends of Intuitionistic Fuzzy Set: A Bibliometric Analysis
- Nurse Scheduling with Opposition-Based Parallel Harmony Search Algorithm
- Some Innovative Types of Fuzzy Ideals in AG-Groupoids
- Extracting Conceptual Relationships and Inducing Concept Lattices from Unstructured Text
- A Hybrid Cuckoo Search and Simulated Annealing Algorithm

