Abstract
Although kallikrein-related peptidase 10 (KLK10) is expressed in a variety of human tissues and body fluids, knowledge of its physiological functions is fragmentary. Similarly, the pathophysiology of KLK10 in cancer is not well understood. In some cancer types, a role as tumor suppressor has been suggested, while in others elevated expression is associated with poor patient prognosis. Active human KLK10 exhibits a unique, three residue longer N-terminus with respect to other serine proteases and an extended 99-loop nearly as long as in tissue kallikrein KLK1. Crystal structures of recombinant ligand-free KLK10 and a Zn2+ bound form explain to some extent the mixed trypsin- and chymotrypsin-like substrate specificity. Zn2+-inhibition of KLK10 appears to be based on a unique mechanism, which involves direct binding and blocking of the catalytic triad. Since the disordered N-terminus and several loops adopt a zymogen-like conformation, the active protease conformation is very likely induced by interaction with the substrate, in particular at the S1 subsite and at the unusual Ser193 as part of the oxyanion hole. The KLK10 structures indicate that the N-terminus, the nearby 75-, 148-, and the 99-loops are connected in an allosteric network, which is present in other trypsin-like serine proteases with several variations.
Introduction
In search for down-regulated genes during breast cancer progression, KLK10 cDNA was originally cloned from radiation-transformed breast epithelial cells and designated normal epithelial cell-specific 1 (NES1) (Liu et al., 1996). KLK10 colocalizes with closely related genes in the chromosomal region 19q13.3–q13.4, forming the family of tissue kallikrein and kallikrein-related peptidases, abbreviated KLKs (Yousef et al., 1999). Gene expression of KLK10 is strongly influenced by estrogens, androgens and progestins (Luo et al., 2000, 2001c, 2003a).
Since both KLK10 mRNA and peptidase expression were lacking in breast and prostate cancer cell lines, a role for KLK10 as tumor suppressor was suggested (Goyal et al., 1998). Downregulation of KLK10 was observed in breast, cervical, prostate, and testicular cancer as well as in acute lymphocytic leukemia (Luo et al., 2001c; Zhang et al., 2006). In line with the potential tumor-suppressing properties, overexpression of KLK10 in prostate cancer cells repressed cell proliferation, while apoptosis increased and glucose metabolism decreased (Hu et al., 2015). However, in other tumor types, such as ovarian, pancreatic, colon, and gastric cancer, upregulation of KLK10 indicates a tumor-supportive role (Yousef et al., 2005). High KLK10 tumor tissue and serum levels are linked with an unfavorable prognosis of ovarian cancer patients (Luo et al., 2001b, 2003b). Upregulation of KLK10 and KLK10 peptidase expression in colorectal and gastric cancer correlate with poor prognosis for patients (Alexopoulou et al., 2013; Jiao et al., 2013). Moreover, many single nucleotide polymorphisms of the KLK10 gene were discovered, as well as alternative transcripts, however, their specific role in physiology and pathology remains unclear (Bharaj et al., 2002; Kurlender et al., 2005; Yousef et al., 2005; Batra et al., 2010).
By using immunofluorometric assays, KLK10 was detected in various human organs and body fluids, such as salivary glands, skin, colon, breast milk, seminal plasma, cerebrospinal fluid, amniotic fluid, and blood serum (Luo et al., 2001a). Further immunohistochemical studies identified KLK10 in many tissues, usually at higher expression levels in adult tissue, in particular in the female reproductive system (Petraki et al., 2002; Shaw and Diamandis, 2007). Enzymatically active KLK10 was identified in ovarian cancer ascites fluid, in which it seems to form complexes with α1-antitrypsin and the inter-α-trypsin inhibitor (Oikonomopoulou et al., 2010). However, the physiological functions of KLK10 are still unclear. Biochemical data on the mutual activation of the KLKs revealed that recombinant KLK10 does not efficiently cleave propeptides of other KLKs, which disfavors a participation in the known KLK activation cascades (Yoon et al., 2007, 2009).
KLK10 is classified in the MEROPS database as serine protease S01.246 of subclan PA(S) (EC 3.4.21.35) as the other 14 members of the human KLK family (Rawlings et al., 2012). According to a standard sequence comparison with (chymo-)trypsin-like enzymes, KLK10 possesses a 33-residue signal peptide, a 13-residue propeptide, with a potentially glycosylated sequon, and a 230-residue catalytic domain, in which it shares 49% identical residues with KLK12 and 46% with KLK8 (Lundwall and Brattsand, 2008; Guo et al., 2014). In contrast to all other KLKs, KLK10 exhibits some unique features on its primary sequence, being the only KLK with a charged N-terminal residue, namely a glutamate in position 16, according to chymotrypsin numbering (Figure 1). All other KLKs possess a hydrophobic residue, such as Ile or Val, which inserts into the activation pocket, in order to form the stabilizing salt bridge to the highly conserved Asp194 (Goettig et al., 2010). Usually, disruption of this salt bridge results in complete loss of activity (Debela et al., 2006a). The unusual N-terminal sequence and lacking protease activity in biological samples, such as ascites fluid from ovarian cancer patients, led to the assumption that it is not a functional serine protease as opposed to the other KLKs (Luo et al., 2006; Zhang et al., 2006).
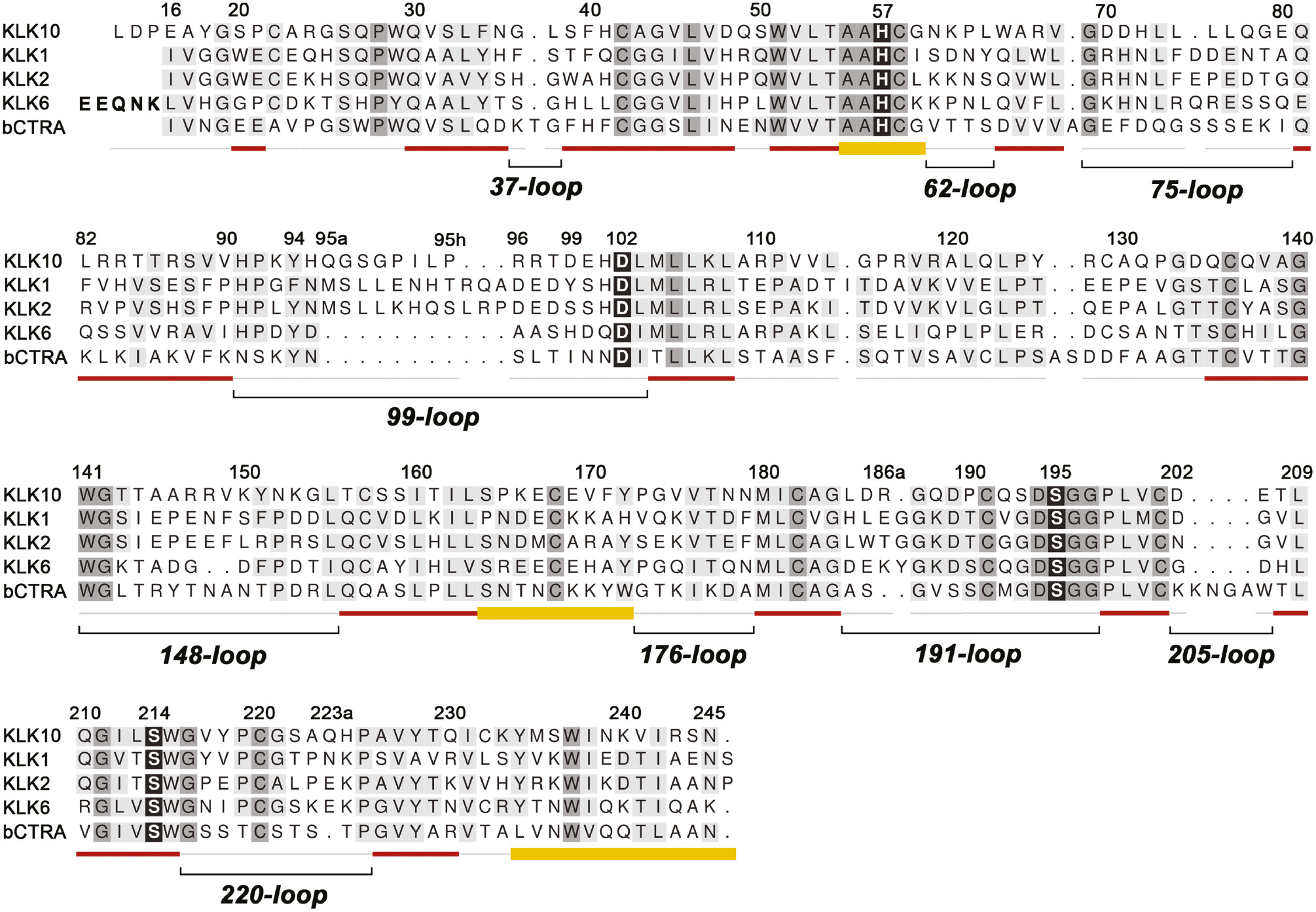
Structure-based alignment of KLK10 with KLKs 1, 2, 6 and bovine chymotrypsin (bCTRA) as numbering reference.
The degree of conservation is indicated by darker background. The catalytic triad residues, and the ‘fourth’ triad residue Ser214 are shown with black background. Secondary structure is depicted as red strands and yellow helices. Loops are shown as light gray lines or, in the case of the relevant surface loops, with additional black lines and names according to the central residue numbering scheme. Insertions with respect to chymotrypsinogen as Gln95A are designated 95a with small letters. The propeptide of KLK6 is indicated with bold letters. Active KLK10 exhibits some uncommon features compared to the other trypsin-like serine proteases, such as Glu16 and the extended N-terminus starting with Leu13. Also the hydrophobic 75-loop segment with four Leu residues is unusual, as well as the 99-loop, which resembles the classical kallikrein loop of KLKs 1, 2 and 3 in length. Instead of a Ser, Thr or Ala, KLK10 possesses the unique Pro190, which may contribute to the mixed tryptic/hydrophobic substrate preference. Finally, Ser193 replaces the highly conserved Gly193 of the oxyanion hole and may considerably influence substrate binding and turnover.
Remarkably, the recombinant E. coli-expressed construct of KLK10 with a three-residue-longer N-terminus starting at Leu13, was active against fluorogenic substrates and yielded a typical specificity profile with a PSSCL library (Debela et al., 2006b). Tryptic specificity could be expected for KLK10, due to the presence of Asp189 at the bottom of the S1 pocket. Intriguingly, the PSSCL measurements demonstrated an unusual dual specificity for the basic P1 residues Lys or Arg, and aliphatic ones, such as Met (Debela et al., 2006b). A similar dual specificity is well established for the kininogen cleavage and inhibitor binding by KLK1 (Müller-Esterl et al., 1986; Chen et al., 2000). Also, the 99-loop comprises an eight-residue insertion, which is nearly as long as the kallikrein loop of the classical KLKs 1, 2 and 3 with their 11-residue insertion. Most unusual and in contrast to all other human KLKs, only KLK10 contains a Ser in position 193, whereas most serine proteases contain the highly conserved Gly193. The backbone amides of Ser195 and Gly193 form the oxyanion pocket, required for stabilization of the tetrahedral intermediate during catalysis (Goettig et al., 2010).
Here, we present the purification, enzymatic characterization, Zn2+ inhibition, and two crystal structures of the long N-terminal form of KLK10, which reveals several unique structural features, such as metal binding in the active site and a novel intermediate state among active KLKs, regarding the N-terminus and loops that surround the active site.
Results and discussion
An extended N-terminus is required for activity of KLK10
The first trials to obtain active human KLK10 with the standard residue number 16, a glutamate at the N-terminus of the mature enzyme were not successful. This finding is not surprising, as all other KLKs possess a hydrophobic residue in this position, typically Ile (nine cases), Leu (3), Val (KLK8), Ala (KLK9). Thus, the first hydrophobic residue before Glu16 was chosen, namely Leu13, which resulted in an active KLK10 peptidase, exhibiting typical activity against fluorogenic and chromogenic substrates, which allowed for substrate specificity profiling from the P4 to P1 positions (Debela et al., 2006b). Taking into account this unusual behavior, it is suggested that Leu13 is designated Leu16 and the following three amino acids Asp16A, Pro16B, and Glu16C.
KLK10 was active against Tos-Gly-Pro-Lys-AMC, however, it seemed to prefer P1-Arg, as corroborated by the following examples of chromogenic and fluorogenic substrates with increasing catalytic efficiency kcat/KM: Z-Gly-Gly-Arg-AMC (2 m−1·s−1), BOC-Phe-Ser-Arg-AMC (95 m−1·s−1), BOC-Val-Pro-Arg-AMC (701 m−1·s−1), and Z-Gly-Pro-Arg-AMC (2076 m−1·s−1) (Debela, 2007). Z and BOC are the carboxybenzyl and tert-butyloxycarbonyl protecting groups of the peptide N-terminus, which correspond to aromatic and aliphatic hydrophobic P4 side chains and may occupy the S4 subsite. By measuring the KLK10 activity against Z-Gly-Pro-Arg-AMC with increasing Zn2+ concentrations, no activity was observed beyond 100 μm. The inhibition constant was determined as IC50=1.6±0.1 μm, which is below the one of KLKs 2–5, 7 and 8 (Goettig et al., 2010).
Overall structure
Despite the presence of the small inhibitor PABA in the crystallization screen, KLK10 crystallized with a ligand-free active site. The corresponding structure will be designated KLK10 and the one that was obtained from crystal soaks with 10 mm ZnCl2 as KLK10-Zn. Both crystals belong to space group P21 and contain two KLK10 molecules per asymmetric unit, with only insignificant deviations between the two independent copies (Figure 2A, B). Already the molecular replacement searches indicated that the target structure of KLK10 differed to some extent from other structures of mature KLKs. The calculation of root mean square deviations using 195 aligned residues resulted for KLK4 (PDB code 2BDG) in a RMSD of 1.63 Å, whereas KLK2 (4NFE) reached 1.43 Å. The zymogen-like conformation of mature KLK3 (3QUM) exhibited an RMSD of 1.47 Å and the only true zymogen structure among the KLKs, namely pro-KLK6 (1GVL), also had a relatively low RMSD with 1.45 Å for 187 aligned residues (Figure 2C). A similarly low RMSD of 1.44 Å was obtained with bovine trypsinogen (2TGT), which eventually was the best basis for a molecular replacement search.
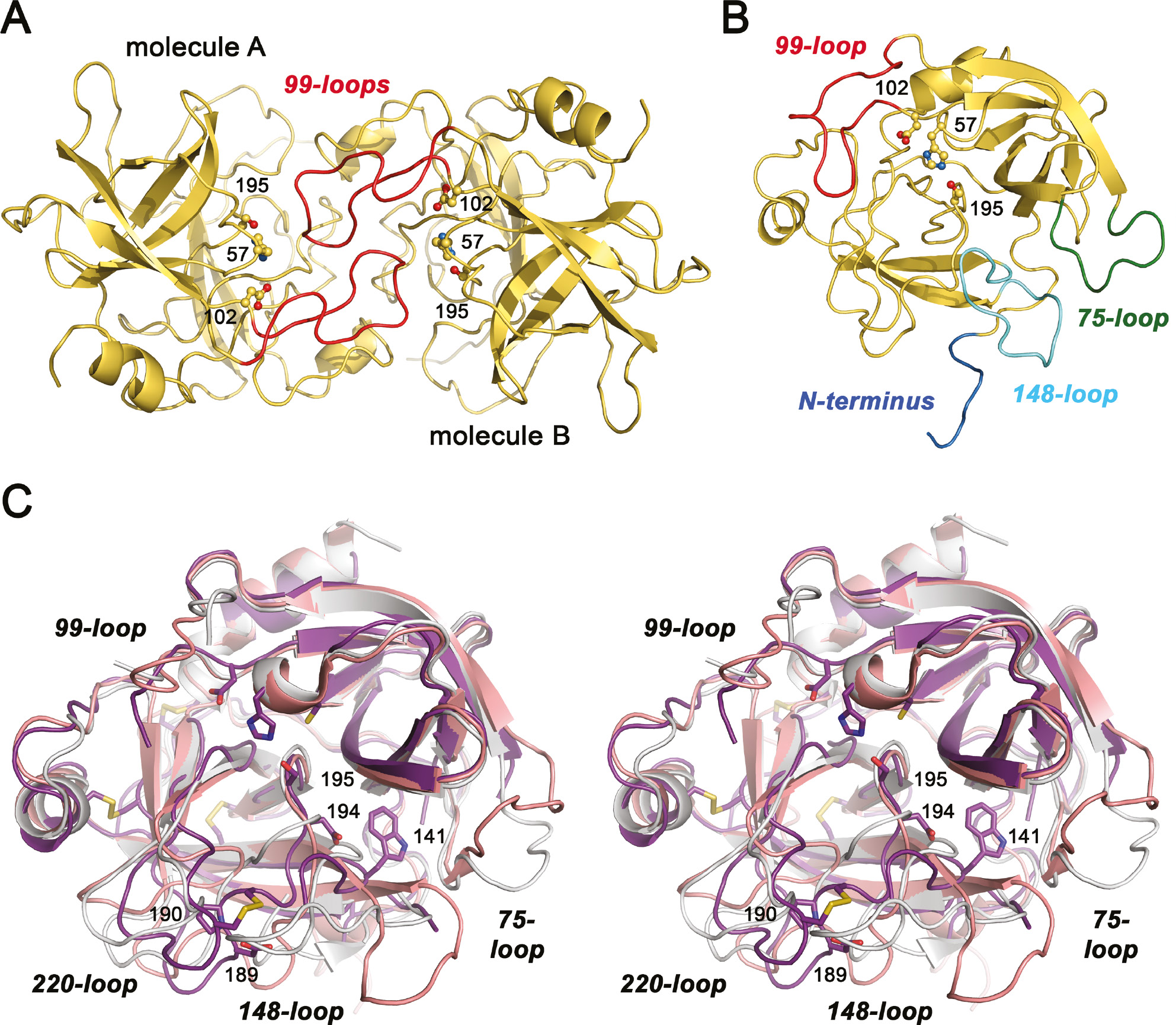
Overall structure of KLK10.
(A) Both KLK10 (yellow) and KLK-Zn (not shown) exhibit a similar dimer in asymmetric unit, with the active sites facing each other. The 99-loops of molecules A and B, shown in red, intercalate to some extent. A stretch of 10 residues from His95 to Arg96 is little defined in the electron density, comprising the whole insertion from Gln95A to Pro95H. The catalytic triad residues Ser195, His57, and Asp102 are depicted as ball-and-stick models and labeled with their sequence numbers. (B) Monomer of KLK10 in standard orientation, with the catalytic triad as ball-and-stick models. In addition to the 99-loop, the other flexible regions of KLK10 are highlighted. The N-terminus (blue) is largely disordered and does not form the activating salt bridge to Asp194. Both the 75-loop (green), which is one residue shorter than most other KLKs, and the 148-loop (aquamarine) lack well-defined electron density for at least six residues. (C) Stereo representation of superposed KLK10-Zn (deep purple), pro-KLK6 (salmon), and active KLK2 (light gray). The disordered regions of KLK10 or of KLK2 have been omitted, e.g. parts of the 99-loop. Catalytic triad residues, the specificity determining Asp189 and Pro190, as well as Asp194, which potentially forms the stabilizing activating salt bridge to the N-terminus, are depicted as stick models and labeled with sequence numbers. Trp141 is also shown, whose backbone amide forms a hydrogen bond to Asp194, as observed for other trypsin-like zymogens. The most relevant loops are labeled regardless of their disorder.
Consequently, the overall fold of KLK10 resembles the prototypic serine protease trypsin, even more its zymogen trypsinogen, with some ingredients from other KLKs (Figure 2B). Active KLK10 consists of 232 amino acid residues, starts with Leu13 (16) and terminates with Asn246. This single polypeptide chain is, as in most other KLKs cross-connected by six disulfide bridges and exhibits the kallikrein-typical cis-Pro219 in the 220-loop (Bode et al., 1983). Otherwise, the structure of KLK10 differs considerably from all other active KLKs, due to extended disordered stretches, starting with the N-terminus and the three major surface loops surrounding the active site (Figure 2B).
The flexible N-terminus and the active site loops of KLK10
Whereas classical KLKs, such as KLK1 or KLK2 mainly exhibit disorder in the 11-residue 99/kallikrein loop, mature KLK10 displays several disordered regions, including the N-terminus, as well as the 75-, 99-, and 148-loops (Laxmikanthan et al., 2005; Skala et al., 2014). Similar to the pro-form of KLK6, the first defined residue in the electron density is Ser20 in chain A and Pro21 in chain B, right before Cys22, which forms a stabilizing disulfide to Cys157 (Gomis-Ruth et al., 2002). This finding confirms that in the present crystal structures the N-terminal residue Leu16 (Leu13 in the common alignment) did not insert into the activation pocket (Figure 3A). In addition, the density around Asp194 does not indicate any salt bridge formation with the α-ammonium group, which would allow to favor the typical rigid conformation of active, mature trypsin-like serine proteases (Bode et al., 1978; Huber and Bode, 1978). Asp194 adopts a similar position as in pro-KLK6 or both bovine trypsinogen and chymotrypsinogen, with a hydrogen bond to the Trp141 backbone amide (Figure 2C). In contrast to pro-KLK6, whose activation pocket is largely occupied by the 191-loop from Lys188 to Gln192, only Gln192 is located in the corresponding relatively open pocket of KLK10 (Figure 3A). These facts alone could classify the present KLK10 variant as zymogen, albeit a rather active one. Zymogen activity is well known for another human serine protease, single-chain tissue type plasminogen activator (sc-tPA), which can adopt an active conformation by insertion of the Lys156 side chain into the activation pocket, with formation of a salt bridge between the Nζ atom and the carboxylate of Asp194 (Renatus et al., 1997). Perhaps Lys150 or Lys153 of KLK10 might fulfill the role of such an activating side chain, but they seem not ideally positioned with respect to the activation pocket.
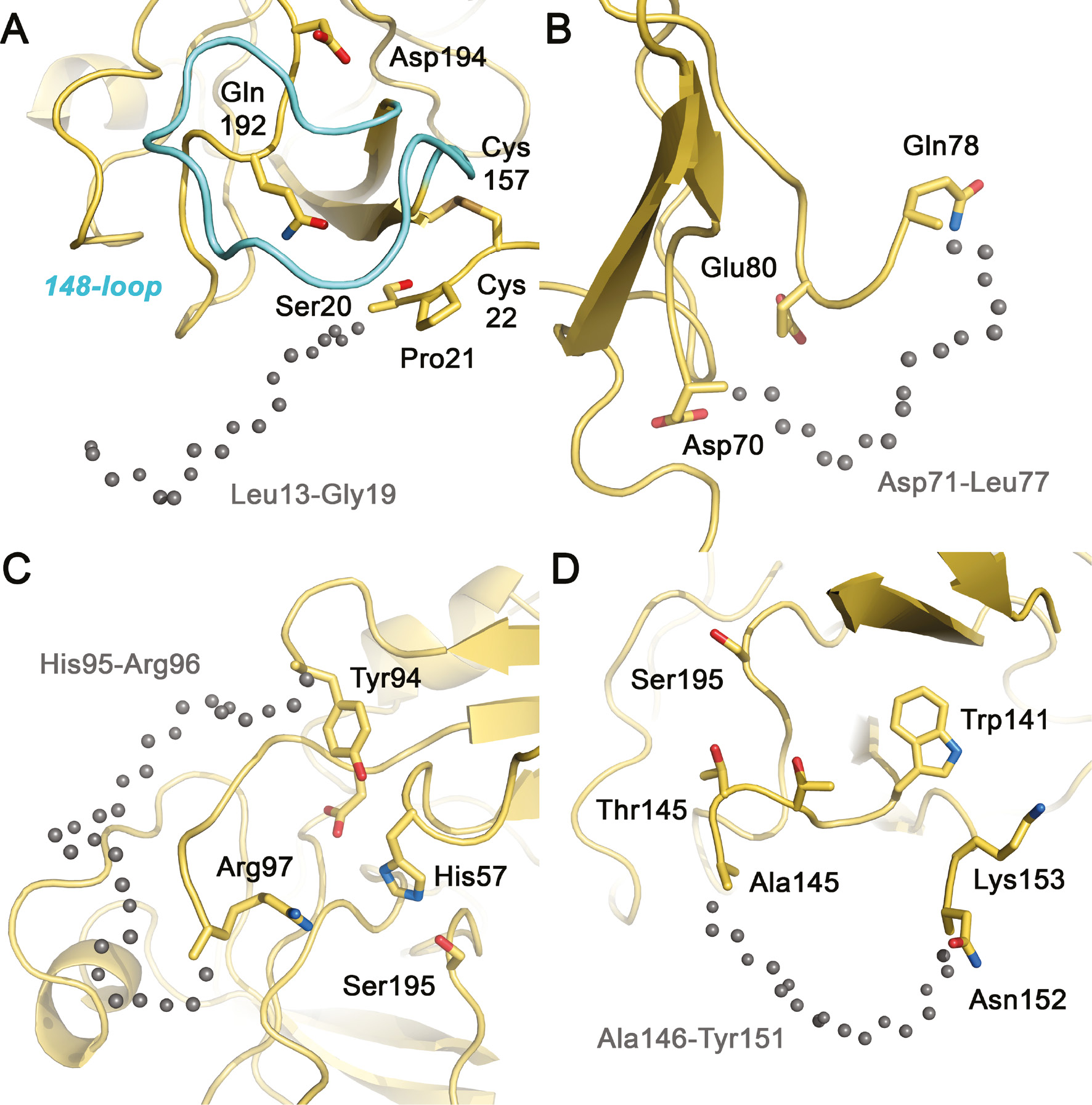
The disordered N-terminus and surface loops of KLK10.
Similar to the same stretches in KLK10-Zn, both molecules A and B lack display disorder without significant electron density in these loops. Gray spheres indicate the likely trace of the disordered main chain segments. (A) The N-terminus of KLK10 is flexible and disordered as in zymogens, e.g. pro-KLK6. Electron density is missing from Leu13 (16) to Gly19, while Ser20 and Pro21 are better defined. (B) The 75-loop exhibits the unusual hydrophobic sequence of four Leu residues, with disorder from Asp71 to Leu77, comprising one deletion with respect to chymotrypsinogen. (C) The 99-loop exhibits the longest disordered stretch of KLK10 from His95 to Arg96, comparable to the same loop of KLK2, which is three residues longer. (D) The 148-loop lacks electron density from Ala146 to Tyr152 in both molecules A, whereas in molecules B the disorder extends to Gly155. Presumably, this loop adopts a wheel-like conformation, which distinguishes it strongly from active conformations and resembles much more the conformation in zymogens.
However, the unusual sequence Leu16-Asp16A-Pro16B-Glu16C does not qualify as typical propeptide either, and may be capable of inserting into the activation pocket in solution, as a series of acetylation experiments of the N-terminus indicates. According to N-terminal sequencing KLK10 possesses a 100% free N-terminus, which upon acetylation is blocked to at least 75%. By contrast, in the presence of the chloromethyl ketone inhibitor PPACK acetylation of KLK10 results in only 25% blocked N-terminus. In addition, the activity against Z-Gly-Pro-Arg-AMC drops to zero turnover for acetylated KLK10, which might represent the trapped zymogen state as present in the crystal structures.
The short 37 and 62-loops of KLK10 do not exhibit significant alterations compared to KLK2 and pro-KLK6, in stark contrast to the larger loops that surround the active site cleft of KLK10. Nevertheless, the massive disorder in the major surface loops of KLK10 is neither typical for a zymogen nor easily explained. For example, bovine chymotrypsinogen (2CGA) is extremely well defined in the electron density, most likely because the N-terminal Cys1 forms a disulfide to Cys122, which is not conserved in the KLKs and appears to suffice in rigidifying the otherwise flexible regions of the activation domain, including the longer loops around the active site and the S1 pocket. The 75-loop of KLK10 with a single residue deletion compared to chymotrypsinogen comprises the unusual sequence DDHLLLLQGE, in which only Asp70 and Glu80 are conserved residues. The electron density in all four copies of KLK10 is missing for Asp71 to Leu77, whereas pro-KLK6 is only disordered from residues 78–80 (Figure 3B). All KLKs, except KLK10, possess mainly charged or polar residues in this loop, which is in general rather well defined and can serve either for Zn2+ or Ca2+ binding, as in most trypsin-like pancreatic/coagulation proteases (Bode and Schwager, 1975; Goettig et al., 2010). For KLK4 an activity regulating allosteric link between the N-terminus and the 75-loop has been observed similar to coagulation factor VIIa (Debela et al., 2006a).
Remarkably, the 99-loop of KLK10 exhibits an eight residue insertion, which is nearly as long as the 11 residue insertion of the three classical KLKs 1, 2 and 3. Seemingly, such long 99-loops can be relatively rigid upon glycosylation on the conserved Asn95 as in KLK1, while the binding of substrates or protein contacts may rigidify it as in KLK3, whereas it is largely disordered in glycan-free KLK2 (Laxmikanthan et al., 2005; Menez et al., 2008; Skala et al., 2014). Since the 99-loop of KLK10 is disordered from His95 to Arg96, including most of the kallikrein loop insertion from Gln95A to Pro95H, its conformation remains hypothetical for this part (Figure 3C). Most of this stretch seems to block the active site, making a wide turn above the active site with the segment Arg97 to Asp99 facing the catalytic His57. Moreover, the 99-loop appears to contribute to the dimer formation of KLK10 in the crystal structure by mutually intercalating 99-loops near the active sites (Figure 2A).
Also, the 148-loop, which is usually well ordered in active trypsin-like proteases, exhibits extended disorder. This loop is known as autolysis loop of the coagulation factors, whose cleavage can lead to inactivation as recently found for KLK2 (Guo et al., 2016). Both molecules A in the KLK10 and KLK10-Zn crystals lack electron density for Ala146 to Tyr151, whereas in molecules B the density is missing for Ala145 to Gly154 (Figure 3D). Again, the corresponding less defined stretch of pro-KLK6 is shorter, ranging only from residues 148–151. However, the disordered segment of bovine trypsinogen extends even from residues 142–151 and beyond the following defined secondary structural elements, comprising the 191- and 220-loops from residues 184–193 and 217–223, respectively.
By contrast, the C-terminal segments of KLK10 are remarkably rigid and only minor disorder occurs in some side chains. Accordingly, the 176-, 191-, 205-, and 220-loops are well defined in the electron density in all four copies of KLK10, which is also independent of the Zn2+, bound in the catalytic center. Essentially, the 176-loop coincides with the conformations of active KLKs, such as KLK2 or KLK4. By contrast, the 191-loop, which shapes large parts of the S1-pocket adopts a zymogen conformation, e.g. with the Asp189 turned towards the molecular surface (Figure 4). Then, the shortened 205-loop displays the expected conformation as in the active KLKs, and also the 220-loop is not too much altered when compared to active KLK2. Due to the disulfide Cys191–Cys220 the 220-loop conformation depends to some extent on the respective conformation of the 191-loop.
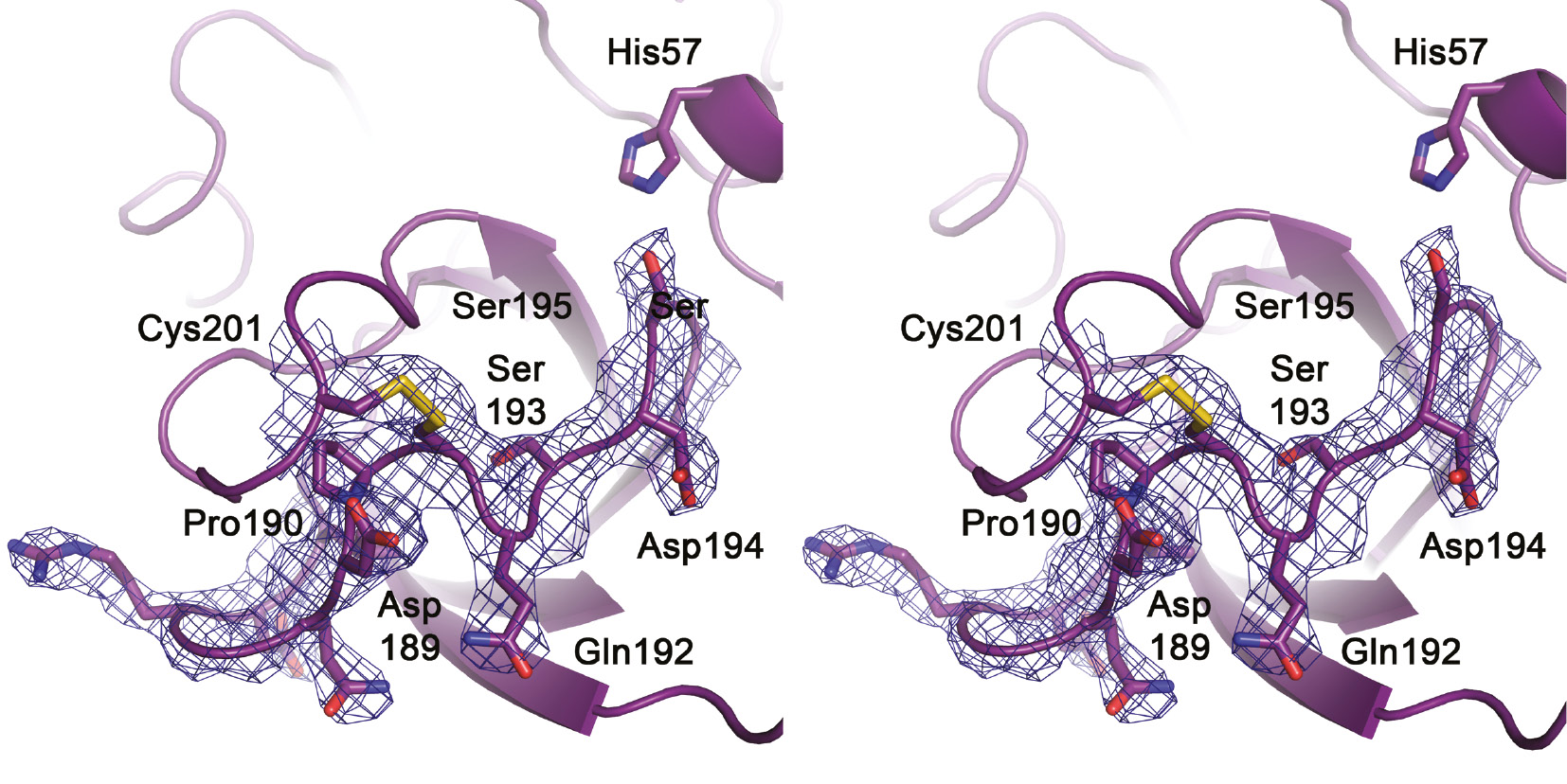
Stereo representation of the 191-loop in KLK10-Zn.
Side chains are represented as sticks and the most relevant residues are labeled, including the catalytic triad residues Ser195 and His57. It is worth noting that the loop is well defined in the 2Fo-Fc electron density (contour 1 σ), similar to the neighbouring 220-loop, to which it is linked by the disulfide Cys191–Cys220. Asp194 is in the typical zymogen-like position turned away from the activation pocket, as in pro-KLK6 or trypsinogen. Nevertheless, the region of the potential salt-bridge formation with the N-terminus near Gln192 is not occluded as in typical zymogens. Ser193 is relocated from the oxyanion hole position, while Asp189 is turned from the bottom of the S1 pocket to the bulk solvent, in contrast to Pro190. This Pro residue might contribute to the mixed tryptic and hydrophobic P1-specificity of KLK10.
Active site and substrate binding pockets
In order to judge the structural characteristics of KLK10 with respect to its active peptidase conformation, the two KLK coordinate data sets with the smallest RMSD are further discussed. As mentioned before, active KLK2 and pro-KLK6 bear the highest similarity to KLK10. Their surface loops share similar conformation or disorder to some extent. Intriguingly, the secondary structure scaffold of all three molecules is essentially the same, even the catalytic triad residues are at their standard position (Figure 2C). However, the catalytic Oγ of Ser195 of molecule A in KLK10-Zn points directly to the Nε2 of His57, while in the other copies it is turned by about 90° towards the S1 pocket as in KLK2, or as in pro-KLK6 it is turned away by 180° with respect to the active protease conformation. Moreover, the backbone amide of Ser193 in KLK10 is relocated from an ideal position as part of the transition state stabilizing oxyanion hole, while its side chain points to the bottom of the S1 pocket without occluding it (Figure 4). Active serine proteases do not require a Gly193 for stabilization of reaction intermediates, as shown by a mutational analysis of coagulation factor XIa elucidating that mainly β-branched residues in place of Gly193 hamper substrate and inhibitor binding (Schmidt et al., 2004, 2008). This finding is corroborated by the example of Phe193 in the kallikrein-like, plasminogen activating snake venom TSV-PA, which does not bind extended polypeptidic substrates and inhibitors (Parry et al., 1998).
Despite the distortion of the KLK10 active site, which depends mainly on the zymogen-like conformations of the N-terminus and the 191-loop, the S1 pocket is relatively wide open and could accommodate a large side chain such as an Arg with little rearrangements (Figure 5). In the current conformation, the primary specificity determining Asp189 is not in the proper position to bind basic side chains. In case the S1 subsite adopts the standard conformation of a serine protease, the Asp189 at its bottom should prefer P1-Arg and P1-Lys residues, in agreement with the profiling results, which show nearly equal acceptance of both basic residues (Debela et al., 2006b). The unusual KLK10 residues, Pro190 and Ala226, which line the distorted S1 pocket, substitute the common Ser190 and Gly226 of KLKs and explain very well that this S1 subsite additionally accepts aliphatic P1 residues, such as Met or probably Cys (Figure 4). The S2 pocket is relatively wide open: only the flexible Arg97 side chain seems to come close to potential P2 residues. Intriguingly, this residue could be the major determinant of the reported preference for Asp and, to a lesser extent, Glu in the S2 subsite (Debela et al., 2006b). The additional preference for Lys and Arg may depend on Asp99 at the back wall of this pocket. Tyr218 is the residue that defines most of the S3 subsite and may be responsible for the overall unspecific character. The slight preference for acidic P3 residues could depend on Arg96 and Arg97, if the little defined, disordered part of the 99-loop extends over the active site cleft. Finally, S4 was very unspecific in profiling experiments and slightly disfavors basic residues, such as P4-His, Lys and especially Arg, while hydrophobic residues rank higher. The S4 subsite is shaped like in other mature KLKs, with a Trp215 as bottom and a Tyr172 as the western wall viewed in standard orientation.
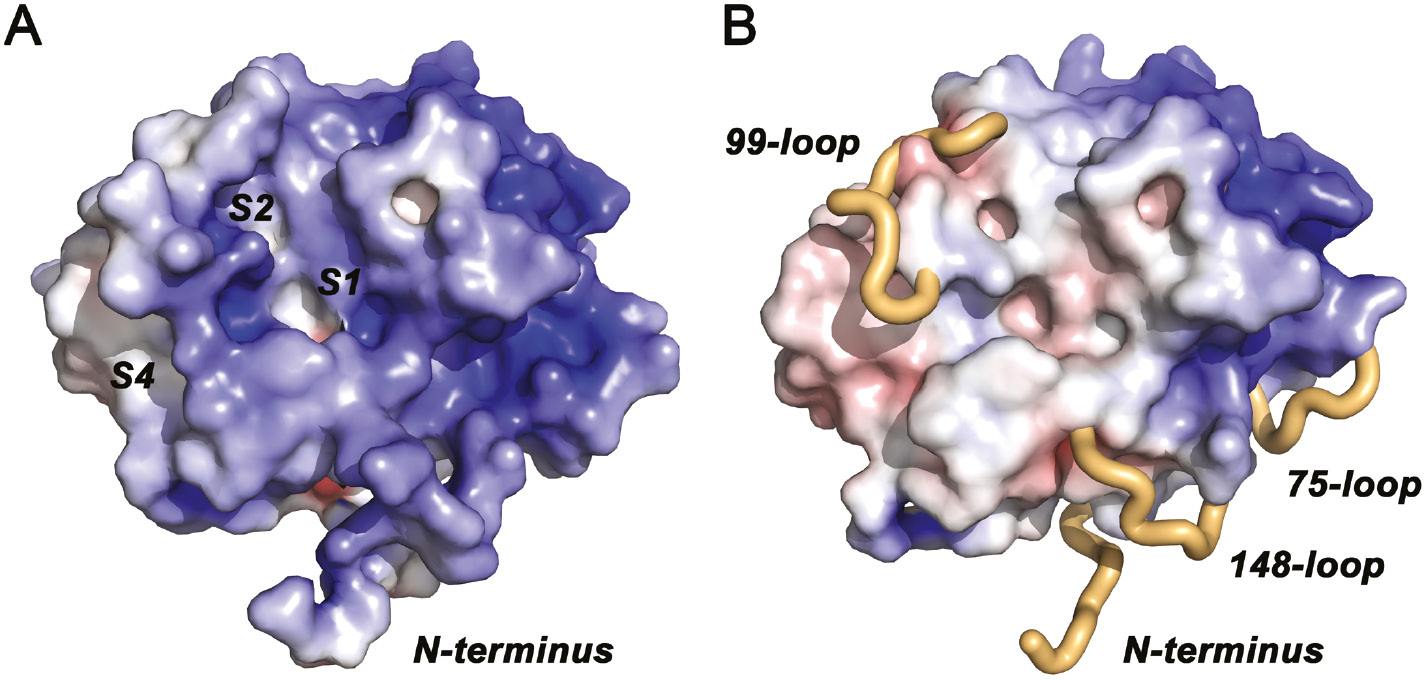
Surface representations of KLK10 with electrostatic potentials.
(A) The electrostatic potential is contoured from −10 kBT/e (red) to +10 kBT/e (blue). Disordered regions were included for the electrostatics calculation and contribute to an overall positive electrostatic potential (KLK10 pI=8.7), which is strongest around the 75-loop. This region corresponds to the anion binding exosite I of thrombin. The specificity pockets S4 to S1 are largely formed as in other active proteases, with S3 partially occupied by the 99-loop and considerable distortions of S1. (B) Disordered regions were omitted for electrostatics calculation and from the molecular surface representation. The flexible N-terminus and the disordered stretches of the 75-, 99-, and 148-loops are depicted as beige tubes.
Zn2+ binding site at the catalytic center
Upon zinc soaking, one site per molecule of the KLK10-dimer becomes visible in the electron density, which is most likely occupied by a Zn2+ ion (Figure 6A). An anomalous Fourier map identified unambiguously an anomalous scatterer at both sites, which confirms the presence of a transition metal. The metal cation is situated on the surface of both KLK10 molecules between the two catalytic triad residues, His57 and Asp102, and is furthermore liganded by Asp99, together forming a trigonal first coordination sphere (Figure 6B). In order to bind this Zn2+, the His57 side chain slightly moves away from Asp102, which also adjusts its side chain towards the metal ion to complete the Zn2+ coordination with Asp99 (Figure 6C). The movement of the imidazolyl side chain of His57 is very subtle: it rotates modestly towards metal cation, not even disconnecting itself from Ser195 (Figure 6D). Average metal ion distances in both KLK10 molecules are 2.1 Å to His57 Nδ1, 1.9 Å to Asp99 Oδ2, and 2.0 Å to Asp102 Oδ2, which is in the range of comparable Zn2+ ligand distances in known structures (Figure 6C). According to the recent literature, the ideal distance of Zn2+ to His is 2.03 Å, and to monodentate Asp 1.99 Å, while bidentate distances range around 2.2 Å (Harding, 2001; Harding et al., 2010).
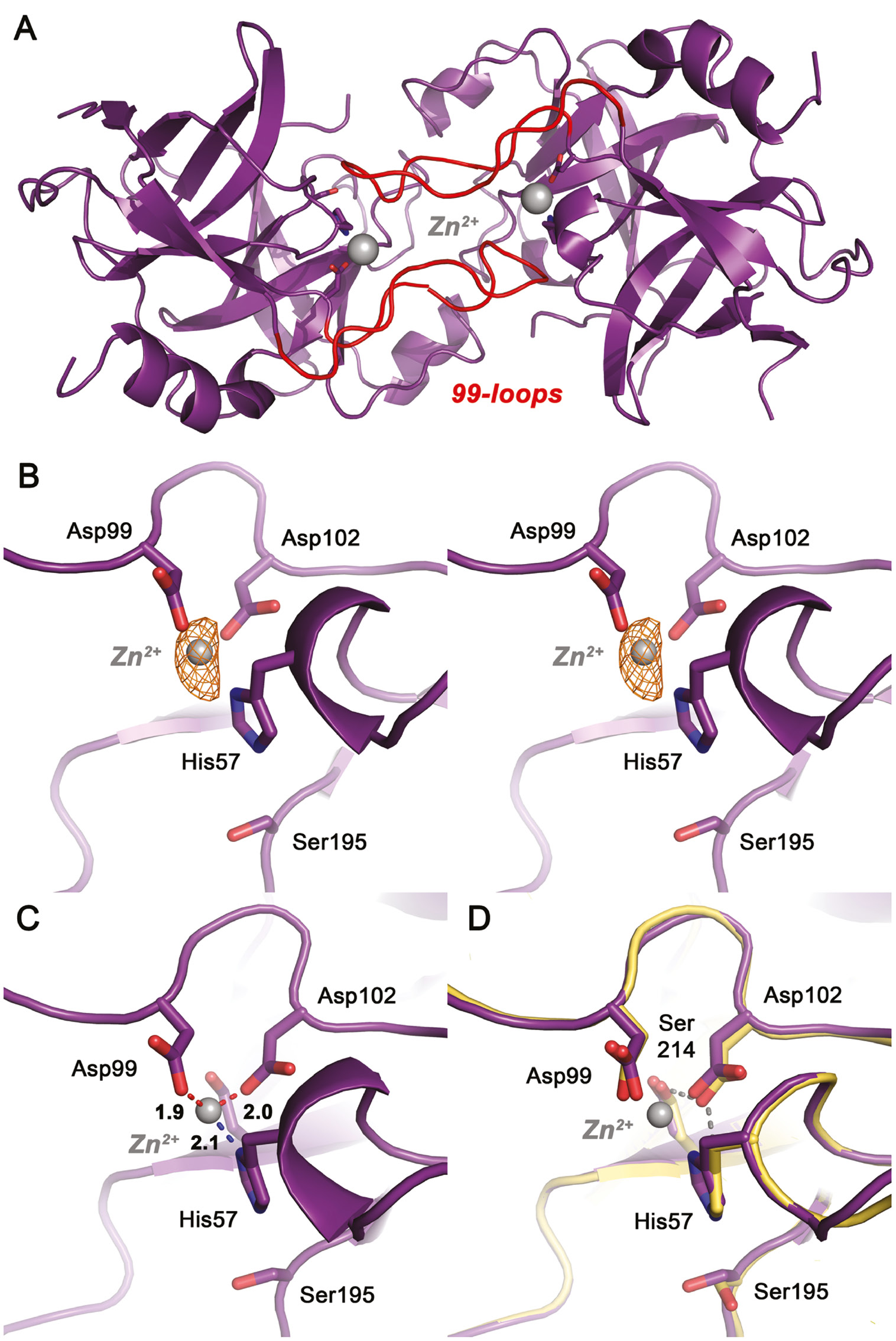
Zn2+ binding to the catalytic center of KLK10-Zn.
(A) Both Zn2+ sites in the KLK10-Zn dimer, with the metal ions displayed as gray sphere. Zn2+ is bound directly in the active site and coordinated by His57, Asp99 and Asp102. The trace of the partially disordered 99-loops is shown in red. (B) Close-up view of the Zn2+ site as stereo representation. The 2Fo-Fc electron density map can be contoured upto 7.0 σ, while two strong anomalous peaks are present, which coincide with Zn2+ sites. An anomalous Fourier map is depicted as orange grid, contoured at 3.5 σ. (C) The trigonal Zn2+ coordination sphere. Average ligand distances of both Zn2+ ions are 2.1 Å for His57, 1.9 Å for Asp99 and 2.0 Å for Asp102. (D) Superposition of the KLK10 Zn2+ structure (deep purple) and Zn2+ free KLK10 (yellow). The catalytic triad undergoes only minor relocations upon Zn2+ binding. The hydrogen bond network of the Zn2+-free structure is depicted as yellow dots. It should be noted that Ser195 and His57 in both strucutures do not exhibit active triad conformations. The Asp102 conformation is stabilized by the highly conserved Ser214, which has been termed the fourth residue of a catalytic tetrad. Zn2+ blocks the triad by interrupting the charge relay system in the region of the hydrogen that is shared by the His57 Nδ1 and the Asp102 Oδ2.
The mechanism of inhibition can be explained by a direct disruption of the catalytic triad, similar to the observations for other KLKs. However, the IC50 of 1.6 μm is the lowest for all investigated Zn2+-inhibited KLKs, which suggests that direct inhibition of the catalytic center is more efficient than for the other examples, which require additional conformational changes such as rotation of His57 out of its catalytic triad position and connection to ligands in the 99-loop (Goettig et al., 2012). For example, the Zn2+-bound structure of KLK10 is different from the crystal structure of the Zn2+-inhibited tonin, where the Zn2+ is liganded by the imidazoyl side chains of His99, His97, and of the active site His57, which has rotated out of its normal site between Ser195 and Asp102, to complete the Zn2+ coordination (Fujinaga and James, 1987). Thus, tonin is more similar to KLK7 and KLK5, which have His99 at the Asp position representing one of the major ligands for Zn2+ binding close to the active site (Debela et al., 2007a,b). However, KLK10 represents the first human serine proteases that holds a metal ion in the core of the active site and involves the two catalytic triad residues, His57 and Asp102. Depending on the respective tissue, the KLK10 activity might be tightly regulated by Zn2+.
Conclusion
In light of all the observations for the KLK10 structures, including the long disordered stretches, and taking into account the relevant comparisons with related structures, it appears that the KLK10 conformation represents neither a typical zymogen nor an active protease. Overall, it resembles less the slow zymogen-like and Na+ activated thrombin form and more coagulation factor VIIa, which exhibits an solvent exposed N-terminus and requires tissue factor as activator (Ruf and Dickinson, 1998; Huntington, 2009). Thus, KLK10 could be considered as sort of a changeling peptidase that is disguised as a zymogen, but ready for immediate protease action, which might be only prevented by the direct binding of Zn2+ to the catalytic triad. Structural and functional evidence from KLK10 are overwhelmingly speaking for the induced-fit model of enzymatic action (Koshland, 1958). However, from a more elevated point of view, this model is no contradiction to the conformational selection model (Ma and Nussinov, 2010). In fact, both models can be reconciled in a very elegant way. In case of the recombinant KLK10 of this study, the variant is simply in a zymogen-like conformation. Most likely, only highly selective substrates can bind to this conformational state and, indeed, induce an active site conformation. In fact, the zymogen-like state exhibits an additional selective element with the Ser193 at the oxyanion pocket. Thus, conformational selection and induced fit are not two sides of the coin, they only represent two extreme views of a single phenomenon (Vogt and Di Cera, 2013). However, KLK10 shows that both positions per se do not suffice. The high flexibility of proteins, whenever it is required, and their persistent rigidity, whenever needed, are intrinsic to KLK10, a unique human protease, with some disordered stretches of high importance for its functionality. In addition to its plasticity, KLK10 activity is regulated in a so far unprecedented manner, by direct metal ion binding and concomitant interruption of the catalytic triad, which appears to be completely independent of the overall conformational state as zymogenic protease.
Regarding the biological and pathophysiological function, we can only present some hypotheses. Despite the wide distribution of KLK10 in many tissues, natural substrates of KLK10 have not yet been characterized, except for a slow cleavage of pro-KLK sequences, which was explained with the presence of the unusual Ser193 (Shaw and Diamandis, 2007; Yoon et al., 2009). Recently, it was demonstrated that pro-KLK3/PSA has considerable zymogen activity, which could extend its functionality, before it is captured by physiological inhibitors (Sangster-Guity et al., 2016). Similarly, KLK10 could remain in a latent, zymogen-like state for long periods of time, without binding inhibitors, such as serpins or α2-macroglobulin.
KLK10 is upregulated in gastric and ovarian cancer, which is associated with a poor prognosis for the patients, however, in breast cancer, it is down-regulated and was proposed as potential tumor suppressor. Similar to other extracellular proteases, such as the metalloproteinases MMP8 and ADAMTS1, or kallikrein-related peptidases, e.g. KLK8, KLK10 might contribute to signaling processes that induce apoptosis or autophagy in cancer cells (Lopez-Otin and Matrisian, 2007). Intriguingly, the eminent tumor suppressor protein p53 is intrinsically disordered, which enables it to bind many diverse interaction partners (Oldfield et al., 2008). Thus, it is tempting to propose that KLK10 recognizes various cancer-related substrates, due to its flexible segments around the active site, in particular the region of the N-terminus, the 75-loop and the 148-loop (Figure 5B). As already mentioned, KLK10 overexpression in prostate cancer cells increases apoptosis and reduces the glucose metabolism (Hu et al., 2015). In addition, the high concentration of Zn2+ in healthy prostate decreases in prostate cancer, while Zn2+ is elevated in breast cancer tissue (Ho and Song, 2009; Riesop et al., 2015). On the one hand, high Zn2+ levels, i.e. above 10 μm, may inhibit the KLK10 protease activity in breast cancer, on the other hand, low Zn2+ concentrations in prostate tumors may increase the KLK10 activity and its tumor-suppressing function.
Materials and methods
Purification and characterization of the active protease
Expression, refolding and chromatographic purification of KLK10 have been described before in detail (Debela et al., 2006b; Debela, 2007). KLK10 was expressed in E. coli M15(pREP4) cells, using a pQE30 vector (Qiagen). Inclusion bodies of KLK10 were dissolved in 8 m urea and the unfolded protein harboring an N-terminal His6-tag was purified via a nickel-nitrilotriacetic acid-Sepharose column. Afterwards, the protein was dialyzed against 4 m urea, 100 mm NaCl, 50 mm Tris/HCl (pH 8.0), and 0.005% Tween-20 for 12 h at room temperature. Refolding took place in 350 mm NaCl, 50 mm Tris/HCl (pH 8.0), 20 mm sucrose, 2 mm CaCl2, 100 mm L-arginine, 5 mm glutathione (reduced), 0.5 mm glutathione (oxidized), 0.005% Tween-20, 0.05% PEG 3350 for 36 h at 4°C. KLK10 samples were further purified or stored in 100 mm NaCl, 50 mm Tris/HCl (pH 8.0) and 0.005% Tween-20, such as during cleavage of the artificial pro-peptide by enterokinase and p-aminobenzamidine-Sepharose affinity chromatography, in which the bound protease was eluted by increasing PABA concentration.
The identity of KLK10 samples was confirmed by N-terminal sequencing. Moreover, the enzymatic activity of KLK10 was initially measured with Z-LR-AMC and Z-FR-AMC, using 20 nm protease in 150 mm NaCl, 50 mm Tris-HCl (pH 8.0), and 0.005% Tween 20 and 1% DMSO at 25°C. The standard substrate concentration was 250 μm and was varied for determining Michaelis-Menten kinetics. For Zn2+ inhibition kinetic measurements with Z-GPR-AMC Tris/HCl buffer (pH 7.0) was employed. Acetylation of the N-terminus was performed with 70 μl samples of 10 nmol KLK10, 18% (v/v) acetic anhydride, 54% (v/v) methanol, 50 mm NH4(HCO3)2 (pH 6.5) for 30 min incubation at room temperature. Afterwards the samples were analyzed by N-terminal sequencing or used in activity assays, respectively.
KLK10 crystallization and data collection
Using the sitting-drop vapor diffusion method, native KLK10 crystals were grown at 18°C from 2 μl protein solution, 10 mg/ml KLK10, which was concentrated from a solution of 30 nm KLK10 with 20 μm PABA, and 2 μl of 100 mm (NH4)2SO4, 100 mm MES (pH 5.6), equilibrated against 500 μl reservoir solution. These crystals belong to the monoclinic space group P21 and contain two molecules per asymmetric unit. For preparation of KLK10-Zn crystals, these crystals were soaked for 2 min in mother liquor with 10 mm ZnCl2. Upon soaking, the crystals remained in monoclinic space group P21 with two molecules per asymmetric, but changed their cell constants to some extent (Table 1). Data were collected at the beamline BW6 of the DESY synchrotron at a wavelength of 1.05 Å (Hamburg, Germany).
Data collection and refinement.
| Data collection | KLK10 | KLK10-Zn |
|---|---|---|
| Space group | P21 | P21 |
| Unit cell dimensions (Å) | a=53.60 b=66.84 c=57.90 | a=52.91 b=67.04 c=57.77 |
| α=90° β=104.29° γ=90° | α=90° β=104.37° γ=90° | |
| Wavelength | 1.05000 | 1.05000 |
| Resolution range (Å) (outer shell) | 51.94–2.70 (2.83–2.70) | 55.96–2.65 (2.78–2.65) |
| Number unique reflections | 10 481 (1355) | 11 339 (1507) |
| Average multiplicity | 3.3 (2.4) | 3.1 (3.1) |
| Rmerge (%) | 5.4 (15.7) | 8.9 (50.6) |
| Rpim (all I+ & I−) | 4.3 (13.4) | 6.7 (38.5) |
| Rano | – | 5.2 (32.0) |
| Completeness (%) | 95.0 (93.3) | 98.7 (99.9) |
| I/σ | 14.4 (4.9) | 8.1 (1.8) |
| CC(1/2) | 0.99 (0.95) | 0.99 (0.72) |
| Refinement | ||
| Resolution in refinement (Å) | 51.94–2.70 (2.80–2.70) | 55.96–2.65 (2.74–2.65) |
| Completeness (%) | 94.4 (90.8) | 98.5 (99.9) |
| Reflections in refinement | 10 400 (982) | 11 317 (1126) |
| Working set/test set | 9812 (921)/588 (61) | 10 724 (1074)/593 (52) |
| Rwork (%)/Rfree (%) | 22.3 (28.2)/28.5 (35.2) | 22.0 (28.1)/27.8 (38.4) |
| Rmsd bond lengths (Å) | 0.011 | 0.011 |
| Rmsd bond angles (°) | 1.128 | 1.433 |
| Protein atoms [B-factor, Å2] | 3107 [31.4] | 3119 [37.4] |
| Ions | 7 SO42− [61.1] | 2 Zn2+ [45.4] 6 SO42− [53.3] |
| Solvent molecules | 135 [26.8] | 124 [36.8] |
| Ramachandran plot | ||
| Most favored regions | 374 (93.6%) | 368 (91.7%) |
| Additionally allowed regions | 26 (6.4%) | 33 (8.3%) |
| Outliers | 0 | 0 |
| PDB codes | 5LPF | 5LPE |
Model building and refinement
The collected diffraction images were integrated with MOSFLM and scaled with AIMLESS of the CCP4 software package (Leslie and Powell, 2007; Winn et al., 2011; Evans and Murshudov, 2013). Molecular replacement search was performed using the KLK4 chain A (PDB code 2BDG) in PHASER and resulted in the following scores: RFZ=9.0, TFZ=7.2, and LLG=+172 and an R-factor of 55.1 (Mccoy et al., 2007). In the following steps, the sequence of KLK10 (O43240, KLK10_HUMAN) from the UniProtKB database was employed (Uniprot, 2015). Using a model comprising 222 residues of bovine trypsinogen (2TGT) improved these values overall to RFZ=8.5, TFZ=7.7, LLG=+320, and an R-factor of 53.1. The correctness of the solution was checked by inspection of proper packing in the asymmetric unit and the unit cell, respectively, and composite omit maps calculated with the CCP4 software. Molecular replacement with the refined solution for the unbound KLK10 coordinates afforded an RFZ=9.6, a final TFZ=27.9, LLG=+3288 and an R-factor of 37.1 for the KLK10-Zn data.
Iterative model building was performed in MAIN and COOT, followed by refinement in PHENIX (Adams et al., 2010; Emsley et al., 2010; Turk, 2013). The polypeptide main chain was overall well defined for all secondary structure elements, while the N-terminus and several surface loops were largely undefined. Rcryst and Rfree of the KLK10 data reached 22.3% and 28.5% at 2.70 Å resolution and for KLK10-Zn 22.0% and 27.8%, at 2.65 Å resolution. For the latter data an anomalous Fourier map was calculated in PHENIX with full model phases and the anomalous differences Δano based on Friedel pairs. More than 90% of the residues of both KLK10 and of KLK10-Zn lies in the favored Ramachandran plot regions and the rest in the allowed regions according to MOLPROBITY (Table 1) (Chen et al., 2010). The coordinates and structure factors were deposited in the protein database under the accession codes 5LPE and 5LPF. RMSD values were calculated using the Gesamt option of the SUPERPOSE software (Krissinel, 2012). All Figures were created with PyMOL (Delano, 2002).
Acknowledgments
We are thankful to Gleb Bourenkov for technical support at the beamline BW6 and to Rainer Friedrich for help with data collection and crystallographic software. P.G. was supported by the Austrian Science Fund (FWF) with project P25003-B21.
References
Adams, P.D., Afonine, P.V., Bunkoczi, G., Chen, V.B., Davis, I.W., Echols, N., Headd, J.J., Hung, L.-W., Kapral, G.J., Grosse-Kunstleve, R.W., et al. (2010). Phenix: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221.10.1107/97809553602060000865Search in Google Scholar
Alexopoulou, D.K., Papadopoulos, I.N., and Scorilas, A. (2013). Clinical significance of kallikrein-related peptidase (KLK10) mRNA expression in colorectal cancer. Clin. Biochem. 46, 1453–1461.10.1016/j.clinbiochem.2013.03.002Search in Google Scholar
Batra, J., Tan, O.L., O’mara, T., Zammit, R., Nagle, C.M., Clements, J.A., Kedda, M.A., and Spurdle, A.B. (2010). Kallikrein-related peptidase 10 (KLK10) expression and single nucleotide polymorphisms in ovarian cancer survival. Int. J. Gynecol. Cancer 20, 529–536.10.1111/IGC.0b013e3181d9273eSearch in Google Scholar
Bharaj, B.B., Luo, L.-Y., Jung, K., Stephan, C., and Diamandis, E.P. (2002). Identification of single nucleotide polymorphisms in the human kallikrein 10 (KLK10) gene and their association with prostate, breast, testicular, and ovarian cancers. Prostate 51, 35–41.10.1002/pros.10076Search in Google Scholar
Bode, W. and Schwager, P. (1975). The single calcium-binding site of crystallin bovin b-trypsin. FEBS Lett. 56, 139–143.10.1016/0014-5793(75)80128-1Search in Google Scholar
Bode, W., Schwager, P., and Huber, R. (1978). The transition of bovine trypsinogen to a trypsin-like state upon strong ligand binding. The refined crystal structures of the bovine trypsinogen-pancreatic trypsin inhibitor complex and of its ternary complex with Ile-Val at 1.9 Å resolution. J. Mol. Biol. 118, 99–112.10.1016/0022-2836(78)90246-2Search in Google Scholar
Bode, W., Chen, Z., Bartels, K., Kutzbach, C., Schmidt-Kastner, G., and Bartunik, H. (1983). Refined 2 a x-ray crystal structure of porcine pancreatic kallikrein a, a specific trypsin-like serine proteinase. Crystallization, structure determination, crystallographic refinement, structure and its comparison with bovine trypsin. J. Mol. Biol. 164, 237–282.10.1016/0022-2836(83)90077-3Search in Google Scholar
Chen, V.C., Chao, L., and Chao, J. (2000). Roles of the p1, p2, and p3 residues in determining inhibitory specificity of kallistatin toward human tissue kallikrein. J. Biol. Chem. 275, 38457–38466.10.1074/jbc.M005605200Search in Google Scholar PubMed
Chen, V.B., Arendall, W.B.R., Headd, J.J., Keedy, D.A., Immormino, R.M., Kapral, G.J., Murray, L.W., Richardson, J.S., and Richardson, D.C. (2010). Molprobity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21.10.1107/97809553602060000884Search in Google Scholar
Debela, M. (2007). Crystal structures of the human tissue kallikreins 4, 5, 7, 10, characterisation of their substrate specificity and analysis of their various zinc inhibition mechanisms. Ph.D. Ph.D. Thesis, TU Munich.Search in Google Scholar
Debela, M., Magdolen, V., Grimminger, V., Sommerhoff, C., Messerschmidt, A., Huber, R., Friedrich, R., Bode, W., and Goettig, P. (2006a). Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site. J. Mol. Biol. 362, 1094–1107.10.1016/j.jmb.2006.08.003Search in Google Scholar PubMed
Debela, M., Magdolen, V., Schechter, N., Valachova, M., Lottspeich, F., Craik, C.S., Choe, Y., Bode, W., and Goettig, P. (2006b). Specificity profiling of seven human tissue kallikreins reveals individual subsite preferences. J. Biol. Chem. 281, 25678–25688.10.1074/jbc.M602372200Search in Google Scholar
Debela, M., Goettig, P., Magdolen, V., Huber, R., Schechter, N.M., and Bode, W. (2007a). Structural basis of the zinc inhibition of human tissue kallikrein 5. J. Mol. Biol. 373, 1017–1031.10.1016/j.jmb.2007.08.042Search in Google Scholar
Debela, M., Hess, P., Magdolen, V., Schechter, N.M., Steiner, T., Huber, R., Bode, W., and Goettig, P. (2007b). Chymotryptic specificity determinants in the 1.0 Å structure of the zinc-inhibited human tissue kallikrein 7. Proc. Natl. Acad. Sci. USA 104, 16086–16091.10.1073/pnas.0707811104Search in Google Scholar
Delano, W.L. (2002). The Pymol Molecular Graphics System (San Carlos, CA, USA: DeLano Scientific).Search in Google Scholar
Emsley, P., Lohkamp, B., Scott, W.G., and Cowtan, K. (2010). Features and development of coot. Acta Crystallogr. D 66, 486–501.10.1107/S0907444910007493Search in Google Scholar
Evans, P.R. and Murshudov, G.N. (2013). How good are my data and what is the resolution? Acta Cryst. D 69, 1204–1214.10.1107/S0907444913000061Search in Google Scholar
Fujinaga, M. and James, M.N. (1987). Rat submaxillary gland serine protease, tonin. Structure solution and refinement at 1.8 Å resolution. J. Mol. Biol. 195, 373–396.10.1016/0022-2836(87)90658-9Search in Google Scholar
Goettig, P., Magdolen, V., and Brandstetter, H. (2010). Natural and synthetic inhibitors of kallikrein-related peptidases (KLKs). Biochimie 92, 1546–1567.10.1016/j.biochi.2010.06.022Search in Google Scholar PubMed PubMed Central
Goettig, P., Debela, M., Bode, W., and Magdolen, V. (2012). Structural aspects of kallikrein-related peptidases. In: Kallikrein-Related Peptidases, V. Magdolen, C.P. Sommerhoff, H. Fritz, and M. Schmitt, eds. (Berlin: De Gruyter), pp. 97–116.10.1515/9783110260373.97Search in Google Scholar
Gomis-Ruth, F.X., Bayes, A., Sotiropoulou, G., Pampalakis, G., Tsetsenis, T., Villegas, V., Aviles, F.X., and Coll, M. (2002). The structure of human prokallikrein 6 reveals a novel activation mechanism for the kallikrein family. J. Biol. Chem. 277, 27273–27281.10.1074/jbc.M201534200Search in Google Scholar PubMed
Goyal, J., Smith, K.M., Cowan, J.M., Wazer, D.E., Lee, S.W., and Band, V. (1998). The role for Nes1 serine protease as a novel tumor suppressor. Cancer Res. 58, 4782–4786.Search in Google Scholar
Guo, S., Skala, W., Magdolen, V., Brandstetter, H., and Goettig, P. (2014). Sweetened kallikrein-related peptidases (KLKs): glycan trees as potential regulators of activation and activity. Biol. Chem. 395, 959–976.10.1515/hsz-2014-0140Search in Google Scholar PubMed
Guo, S., Skala, W., Magdolen, V., Briza, P., Biniossek, M.L., Schilling, O., Kellermann, J., Brandstetter, H., and Goettig, P. (2016). A single glycan at the 99-loop of human kallikrein-related peptidase 2 regulates activation and enzymatic activity. J. Biol. Chem. 291, 593–604.10.1074/jbc.M115.691097Search in Google Scholar PubMed PubMed Central
Harding, M. (2001). Geometry of metal-ligand interactions in proteins. Acta Crystallogr. D 57, 401–411.10.1107/S0907444900019168Search in Google Scholar PubMed
Harding, M., Nowicki, M.W., and Walkinshaw, M.D. (2010). Metals in protein structures: a review of their principal features. Crystallogr. Rev. 16, 247–302.10.1080/0889311X.2010.485616Search in Google Scholar
Ho, E. and Song, Y. (2009). Zinc and prostatic cancer. Curr. Opin. Clin. Nutr. Metab. Care 12, 640–645.10.1097/MCO.0b013e32833106eeSearch in Google Scholar PubMed PubMed Central
Hu, J., Lei, H., Fei, X., Liang, S., Xu, H., Qin, D., Wang, Y., Wu, Y., and Li, B. (2015). Nes1/KLK10 gene represses proliferation, enhances apoptosis and down-regulates glucose metabolism of pc3 prostate cancer cells. Sci. Rep. 5, 1–14.10.1038/srep17426Search in Google Scholar PubMed PubMed Central
Huber, R. and Bode, W. (1978). Structural basis of the activation and action of trypsin. Acc. Chem. Res. 11, 114–122.10.1021/ar50123a006Search in Google Scholar
Huntington, J.A. (2009). Slow thrombin is zymogen-like. J. Thromb. Haemost. 7, 159–164.10.1111/j.1538-7836.2009.03365.xSearch in Google Scholar PubMed PubMed Central
Jiao, X., Lu, H.J., Zhai, M.M., Tan, Z.J., Zhi, H.N., Liu, X.M., Liu, C.H., and Zhang, D.P. (2013). Overexpression of kallikrein gene 10 is a biomarker for predicting poor prognosis in gastric cancer. World J. Gastroenterol. 19, 9425–9431.10.3748/wjg.v19.i48.9425Search in Google Scholar PubMed PubMed Central
Koshland, J.D.E. (1958). Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. USA 44, 98–104.10.1073/pnas.44.2.98Search in Google Scholar PubMed PubMed Central
Krissinel, E. (2012). Enhanced fold recognition using efficient short fragment clustering. J. Mol. Biochem. 1, 76–85.Search in Google Scholar
Kurlender, L., Borgono, C., Michael, I.P., Obiezu, C., Elliott, M.B., Yousef, G.M., and Diamandis, E.P. (2005). A survey of alternative transcripts of human tissue kallikrein genes. Biochim. Biophys. Acta 1755, 1–14.10.1016/j.bbcan.2005.02.001Search in Google Scholar PubMed
Laxmikanthan, G., Blaber, S.I., Bernett, M.J., Scarisbrick, I.A., Juliano, M.A., and Blaber, M. (2005). 1.70 Å X-ray structure of human apo kallikrein 1: structural changes upon peptide inhibitor/substrate binding. Proteins 58, 802–814.10.1002/prot.20368Search in Google Scholar PubMed
Leslie, A.G.W. and Powell, H.R. (2007). Processing diffraction data with mosflm. In: Evolving methods for macromolecular crystallography, R.J. Read and J. Sussman, eds. (Netherlands: Springer), pp. 41–51.10.1007/978-1-4020-6316-9_4Search in Google Scholar
Liu, X.L., Wazer, D.E., Watanabe, K., and Band, V. (1996). Identification of a novel serine protease-like gene, the expression of which is down-regulated during breast cancer progression. Cancer Res. 56, 3371–3379.Search in Google Scholar
Lopez-Otin, C. and Matrisian, L.M. (2007). Emerging roles of proteases in tumour suppression. Nat. Rev. Cancer 7, 800–808.10.1038/nrc2228Search in Google Scholar PubMed
Lundwall, A. and Brattsand, M. (2008). Kallikrein-related peptidases. Cell. Mol. Life Sci. 65, 2019–2038.10.1007/s00018-008-8024-3Search in Google Scholar PubMed
Luo, L.Y., Grass, L., and Diamandis, E.P. (2000). The normal epithelial cell-specific 1 (Nes1) gene is up-regulated by steroid hormones in the breast carcinoma cell line bt-474. Anticancer Res. 20, 981–986.Search in Google Scholar
Luo, L.Y., Grass, L., Howarth, D.J.C., Thibault, P., Ong, H., and Diamandis, E.P. (2001a). Immunofluorometric assay of human kallikrein 10 and its identification in biological fluids and tissues. Clin. Chem. 47, 237–246.10.1093/clinchem/47.2.237Search in Google Scholar
Luo, L.Y., Katsaros, D., Scorilas, A., Fracchioli, S., Piccinno, R., Rigault De La Longrais, I.A., Howarth, D.J.C., and Diamandis, E.P. (2001b). Prognostic value of human kallikrein 10 expression in epithelial ovarian carcinoma. Clin. Cancer Res. 7, 2372–2379.Search in Google Scholar
Luo, L.Y., Meyts, E.R.-D., Jung, K., and Diamandis, E.P. (2001c). Expression of the normal epithelial cell-specific 1 (Nes1; KLK10) candidate tumour suppressor gene in normal and malignant testicular tissue. Br. J. Cancer 85, 220–224.10.1054/bjoc.2001.1870Search in Google Scholar PubMed PubMed Central
Luo, L.Y., Grass, L., and Diamandis, E.P. (2003a). Steroid hormone regulation of the human kallikrein 10 (KLK10) gene in cancer cell lines and functional characterization of the KLK10 gene promoter. Clin. Chim. Acta 337, 115–126.10.1016/j.cccn.2003.07.008Search in Google Scholar PubMed
Luo, L.Y., Katsaros, D., Scorilas, A., Fracchioli, S., Bellino, R., Van Gramberen, M., De Bruijn, H., Henrik, A., Stenman, U.-H., Massobrio, M., et al. (2003b). The serum concentration of human kallikrein 10 represents a novel biomarker for ovarian cancer diagnosis and prognosis. Cancer Res. 63, 807–811.Search in Google Scholar
Luo, L.Y., Soosaipillai, A., Grass, L., and Diamandis, E.P. (2006). Characterization of human kallikreins 6 and 10 in ascites fluid from ovarian cancer patients. Tumour Biol. 27, 227–234.10.1159/000094693Search in Google Scholar PubMed
Ma, B. and Nussinov, R. (2010). Enzyme dynamics point to stepwise conformational selection in catalysis. Curr. Opin. Chem. Biol. 14, 652–659.10.1016/j.cbpa.2010.08.012Search in Google Scholar PubMed PubMed Central
Mccoy, A.J., Grosse-Kunstleve, R.W., Adams, P.D., Winn, M.D., Storoni, L.C., and Read, R.J. (2007). Phaser crystallographic software. J. Appl. Cryst. 40, 658–674.10.1107/S0021889807021206Search in Google Scholar PubMed PubMed Central
Menez, R., Michel, S., Muller, B.H., Bossus, M., Ducancel, F., Jolivet-Reynaud, C., and Stura, E.A. (2008). Crystal structure of a ternary complex between human prostate-specific antigen, its substrate acyl intermediate and an activating antibody. J. Mol. Biol. 376, 1021–1033.10.1016/j.jmb.2007.11.052Search in Google Scholar
Müller-Esterl, W., Iwanaga, S., and Nakanishi, S. (1986). Kininogens revisited. Trends Biochem. Sci. 11, 336–339.10.1016/0968-0004(86)90293-8Search in Google Scholar
Oikonomopoulou, K., Batruch, I., Smith, C.R., Soosaipillai, A., Diamandis, E.P., and Hollenberg, M.D. (2010). Functional proteomics of kallikrein-related peptidases in ovarian cancer ascites fluid. Biol. Chem. 391, 381–390.10.1515/bc.2010.045Search in Google Scholar
Oldfield, C.J., Meng, J., Yang, J.Y., Yang, M.Q., Uversky, V.N., and Dunker, A.K. (2008). Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 9, 1–20.10.1186/1471-2164-9-S1-S1Search in Google Scholar
Parry, M.a.A., Jacob, U., Huber, R., Wisner, A., Bon, C., and Bode, W. (1998). The crystal structure of the novel snake venom plasminogen activator TSV-PA: a prototype structure for snake venom serine proteinases. Structure 6, 1195–1206.10.1016/S0969-2126(98)00119-1Search in Google Scholar
Petraki, C.D., Karavana, V.N., Luo, L.-Y., and Diamandis, E.P. (2002). Human kallikrein 10 expression in normal tissues by immunohistochemistry. J. Histochem. Cytochem. 50, 1247–1261.10.1177/002215540205000912Search in Google Scholar
Rawlings, N.D., Barrett, A.J., and Bateman, A. (2012). Merops: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–350.10.1093/nar/gkr987Search in Google Scholar
Renatus, M., Engh, R.A., Stubbs, M.T., Huber, R., Fischer, S., Kohnert, U., and Bode, W. (1997). Lysine 156 promotes the anomalous proenzyme activity of tpa: X-ray crystal structure of single-chain human TPA. EMBO J. 16, 4797–4805.10.1093/emboj/16.16.4797Search in Google Scholar
Riesop, D., Hirner, A.V., Rusch, P., and Bankfalvi, A. (2015). Zinc distribution within breast cancer tissue: a possible marker for histological grading? J. Cancer Res. Clin. Oncol. 141, 1321–1331.10.1007/s00432-015-1932-3Search in Google Scholar
Ruf, W. and Dickinson, C.D. (1998). Allosteric regulation of the cofactor-dependent serine protease coagulation factor VIIa. Trends Cardiovasc. Med. 8, 350–356.10.1016/S1050-1738(98)00031-0Search in Google Scholar
Sangster-Guity, N., Tu-Sekine, B., Raben, D.M., Denmeade, S.R., and Williams, S.A. (2016). Mutational analysis of prostate-specific antigen defines the intrinsic proteolytic activity of the proPSA zymogen. Prostate 76, 1203–1217.10.1002/pros.23216Search in Google Scholar PubMed
Schmidt, A.E., Ogawa, T., Gailani, D., and Bajaj, S.P. (2004). Structural role of Gly193 in serine proteases. J. Biol. Chem. 279, 29485–29492.10.1074/jbc.M402971200Search in Google Scholar PubMed
Schmidt, A.E., Sun, M.-F., Ogawa, T., Bajaj, S.P., and Gailani, D. (2008). Functional role of residue 193 (chymotrypsin numbering) in serine proteases: influence of side chain length and b-branching on the catalytic activity of blood coagulation factor XIa. Biochemistry 47, 1326–1335.10.1021/bi701594jSearch in Google Scholar PubMed PubMed Central
Shaw, J.L.V. and Diamandis, E.P. (2007). Distribution of 15 human kallikreins in tissues and biological fluids. Clin. Chem. 53, 1423–1432.10.1373/clinchem.2007.088104Search in Google Scholar PubMed
Skala, W., Utzschneider, D.T., Magdolen, V., Debela, M., Guo, S., Craik, C.S., Brandstetter, H., and Goettig, P. (2014). Structure-function analyses of human kallikrein-related peptidase 2 establish the 99-loop as master regulator of activity. J. Biol. Chem. 289, 34267–34283.10.1074/jbc.M114.598201Search in Google Scholar PubMed PubMed Central
Turk, D. (2013). Main software for density averaging, model building, structure refinement and validation. Acta Crystallogr. D 69, 1342–1357.10.1107/S0907444913008408Search in Google Scholar PubMed PubMed Central
Uniprot, C. (2015). Uniprot: a hub for protein information. Nucleic Acids Res. 43, D204–D212.10.1093/nar/gku989Search in Google Scholar PubMed PubMed Central
Vogt, A.D. and Di Cera, E. (2013). Conformational selection is a dominant mechanism of ligand binding. Biochemistry 52, 5723–5729.10.1021/bi400929bSearch in Google Scholar PubMed PubMed Central
Winn, M.D., Ballard, C.C., Cowtan, K.D., Dodson, E.J., Emsley, P., Evans, P.R., Keegan, R.M., Krissinel, E.B., Leslie, A.G.W., Mccoy, A., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242.10.1107/S0907444910045749Search in Google Scholar PubMed PubMed Central
Yoon, H., Laxmikanthan, G., Lee, J., Blaber, S.I., Rodriguez, A., Kogot, J.M., Scarisbrick, I.A., and Blaber, M. (2007). Activation profiles and regulatory cascades of the human kallikrein-related peptidases. J. Biol. Chem. 282, 31852–31864.10.1074/jbc.M705190200Search in Google Scholar PubMed
Yoon, H., Blaber, S.I., Debela, M., Goettig, P., Scarisbrick, I.A., and Blaber, M. (2009). A completed KLK activome profile: investigation of activation profiles of KLK9, 10, and 15. Biol. Chem. 390, 373–377.10.1515/BC.2009.026Search in Google Scholar PubMed PubMed Central
Yousef, G.M., Luo, L.Y., and Diamandis, E.P. (1999). Identification of novel human kallikrein-like genes on chromosome 19q13. 3–q13.4. Anticancer Res. 19, 2843–2852.Search in Google Scholar
Yousef, G.M., White, N.M., Michael, I.P., Cho, J.C., Robb, J.D., Kurlender, L., Khan, S., and Diamandis, E.P. (2005). Identification of new splice variants and differential expression of the human kallikrein 10 gene, a candidate cancer biomarker. Tumour Biol. 26, 227–235.10.1159/000087377Search in Google Scholar PubMed
Zhang, Y., Bhat, I., Zeng, M., Jayal, G., Wazer, D.E., Band, H., and Band, V. (2006). Human kallikrein 10, a predictive marker for breast cancer. Biol. Chem. 387, 715–721.10.1515/BC.2006.090Search in Google Scholar PubMed
©2016, Peter Goettig et al., published by De Gruyter
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 3.0 License.
Articles in the same Issue
- Frontmatter
- Guest Editorial
- Highlight: remodelling the KLK landscape down under
- HIGHLIGHT: 6TH INTERNATIONAL SYMPOSIUM ON KALLIKREINS AND KALLIKREIN-RELATED PEPTIDASES
- Kallikrein(K1)-kinin-kininase (ACE) and end-organ damage in ischemia and diabetes: therapeutic implications
- Mechanistic insight from murine models of Netherton syndrome
- Development of molecules stimulating the activity of KLK3 – an update
- Exploring the active site binding specificity of kallikrein-related peptidase 5 (KLK5) guides the design of new peptide substrates and inhibitors
- Structural basis for the Zn2+ inhibition of the zymogen-like kallikrein-related peptidase 10
- Clinical relevance of kallikrein-related peptidase 6 (KLK6) and 8 (KLK8) mRNA expression in advanced serous ovarian cancer
- Kallikrein-related peptidase 6 exacerbates disease in an autoimmune model of multiple sclerosis
- A viable mouse model for Netherton syndrome based on mosaic inactivation of the Spink5 gene
- Therapeutic modulation of tissue kallikrein expression
- In vitro evidence that KLK14 regulates the components of the HGF/Met axis, pro-HGF and HGF-activator inhibitor 1A and 1B
- A computational analysis of the genetic and transcript diversity at the kallikrein locus
- Reviews
- Lymphocyte signaling and activation by the CARMA1-BCL10-MALT1 signalosome
- The power, pitfalls and potential of the nanodisc system for NMR-based studies
- Research Articles/Short Communications
- Cell Biology and Signaling
- Synergistic induction of cardiomyocyte differentiation from human bone marrow mesenchymal stem cells by interleukin 1β and 5-azacytidine
Articles in the same Issue
- Frontmatter
- Guest Editorial
- Highlight: remodelling the KLK landscape down under
- HIGHLIGHT: 6TH INTERNATIONAL SYMPOSIUM ON KALLIKREINS AND KALLIKREIN-RELATED PEPTIDASES
- Kallikrein(K1)-kinin-kininase (ACE) and end-organ damage in ischemia and diabetes: therapeutic implications
- Mechanistic insight from murine models of Netherton syndrome
- Development of molecules stimulating the activity of KLK3 – an update
- Exploring the active site binding specificity of kallikrein-related peptidase 5 (KLK5) guides the design of new peptide substrates and inhibitors
- Structural basis for the Zn2+ inhibition of the zymogen-like kallikrein-related peptidase 10
- Clinical relevance of kallikrein-related peptidase 6 (KLK6) and 8 (KLK8) mRNA expression in advanced serous ovarian cancer
- Kallikrein-related peptidase 6 exacerbates disease in an autoimmune model of multiple sclerosis
- A viable mouse model for Netherton syndrome based on mosaic inactivation of the Spink5 gene
- Therapeutic modulation of tissue kallikrein expression
- In vitro evidence that KLK14 regulates the components of the HGF/Met axis, pro-HGF and HGF-activator inhibitor 1A and 1B
- A computational analysis of the genetic and transcript diversity at the kallikrein locus
- Reviews
- Lymphocyte signaling and activation by the CARMA1-BCL10-MALT1 signalosome
- The power, pitfalls and potential of the nanodisc system for NMR-based studies
- Research Articles/Short Communications
- Cell Biology and Signaling
- Synergistic induction of cardiomyocyte differentiation from human bone marrow mesenchymal stem cells by interleukin 1β and 5-azacytidine

