Abstract
4-(2′,4′-Difluorobiphenyl-4-yl)-6-arylpyrimidin-2-amines 5a–g were synthesized and evaluated for their ability to inhibit various bacterial and fungal strains. The cytotoxicity of the compounds was investigated by a 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay against the Hep-2 cell line. Molecular docking studies were also conducted to locate the binding interaction with the target protein 4LRH.
Introduction
Pyrimidine is a key structural moiety of natural products, such as vitamins, coenzymes, and uric acid, as well as of many synthetic drugs including veronal, sulfadiazine, fluorouracil, glivec, and rosuvastatin [1], [2]. DNA and RNA contain pyrimidine fragments [3]. A wide range of biological activities have been noted for pyrimidine-containing natural products and synthetic molecules [4], [5], [6], [7], [8]. In particular, 2-aminopyrimidine derivatives have found important applications in pharmacology and as synthetic precursors in medicinal chemistry [9], [10]. Many fluorine-containing organic compounds including fluorinated pyrimidines are known to exhibit a broad range of biological activities [11], [12], [13], [14]. Herein we report the synthesis and biological evaluation of new 4-(2′,4′-difluorobiphenyl-4-yl)-6-arylpyrimidin-2-amines 5a–g.
Results and discussion
Chemistry
The desired compounds 5a–g were synthesized as shown in Scheme 1 using the synthetic methodology described previously for similar derivatives [15], [16]. The key intermediates 3a–g were prepared by the Claisen-Schmidt condensation method as reported previously [17]. The structures of the synthesized compounds were characterized by elemental analysis, IR, 1H NMR, 13C NMR spectroscopy, and mass spectrometry. The spectral analysis is fully consistent with the given structures. In several cases, the tentative assignments made by analysis of 1H NMR and 13C NMR spectra were verified by analysis of the 1H-13C COSY spectra. The electrospray ionization mass spectra of all products show the expected peak at [M+1]+.

Antimicrobial evaluation
Compounds 5a–g were tested against Gram-positive bacteria (Bacillus subtilis and Staphylococcus aureus), Gram-negative bacteria (Pseudomonas aeruginosa, Escherichia coli, and Klebsiella pneumoniae), and fungi (Candida albicans) by the disc diffusion method. Additional evaluation was carried out on compounds 5a–g to determine their minimum inhibitory concentration (MIC) values using the two-fold serial dilution method. The results are illustrated in Figures 1 and 2 (Tables S1 and S2 in the Supplementary Material). The mean zone of inhibition produced by these compounds against tested bacterial and fungal strains ranged from 7 mm to 16 mm. Under similar conditions, gentamycin, used as a positive control, produced a mean zone of inhibition ranging between 17 mm and 20 mm. It can be seen that the fluorine-substituted derivative 5b possesses 75%–80% of the activity of gentamycin against all bacterial strains. Compound 5g with methoxy substitution at the ortho position exhibits superior activity towards the fungal strain C. albicans. The MIC results support the conclusion above that compound 5b has a pronounced antibacterial activity. All other derivatives, 5a and 5c–g, exert only moderate activities against the tested organisms.

Antimicrobial activity of compounds 5a–g obtained by disc diffusion method.

Antimicrobial activity of compounds 5a–g obtained by two-fold serial dilution method.
Cytotoxicity
Compounds 5a–g were assayed for their in vitro cytotoxic activity against the Hep-2 laryngeal carcinoma cell line by the 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay with maintaining a control (with solvent only). The results are shown in Figure 3. In general, the pyrimidine derivatives exhibit good cytotoxic activity against various cell lines like melanoma, colon cancer, ovarian cancer, and breast cancer [18]. The results also indicate that the compounds are strongly inhibitory against the Hep-2 cell line. Based on the substitution on the phenyl ring, the cytotoxic activity decreases in the order of 4-F>H>2-OCH3>4-Br>4-Cl>4-CH3>3-NO2. It is evident that compound 5b with lipophilic fluorine substitution exhibits the most potent inhibitory activity with a cytotoxicity of 70.5%. The results are in agreement with previous findings that incorporation of fluorine enhances the pharmacological activity of the modified molecule [19].

Cell viability of compounds 5a–g.
Molecular docking studies
Molecular docking is a promising tool to gain insight into the binding of the ligand and protein. Docking studies were made with protein carotenoid dehydrosqualene synthase from S. aureus complexed with BPH-673 (3ACX) and human folate receptor alpha in complex with folic acid (4LRH). Docking results of the ligands were expressed as a G score (Table 1), which is an empirical function that includes many factors like hydrophobicity, hydrogen bonding, and rotation penalty, among others. The docked ligands were ranked based on various types of interactions between the ligand and protein. The results reveal that all ligands can accommodate the active pocket of the receptor and show an excellent G score ranging from –10.9 to –9.9. The 3D- and 2D-binding modes of the best ligand 5g are shown in Figure 4.
Docking scores of compounds 5a–g.
| Compound | 5a | 5b | 5c | 5d | 5e | 5f | 5g |
|---|---|---|---|---|---|---|---|
| G score | –10.0 | –9.9 | –9.9 | –9.9 | –10.0 | –9.9 | –10.9 |
| Interacting residues | TRP 171, TRP 140, HIE 135, ARG 103 | TRP 171, TYR 60, TYR 85, THR 82 | TRP 171, TRP 140, HIE 135, ARG 103 | TRP 171, TRP 140, HIE 135, ARG 103 | TRP 171, TRP 140, HIE 135, ARG 103 | TRP 171, TRP 140, HIE 135, ARG 103 | TRP 171, TRP 140, HIE 135, H2O |
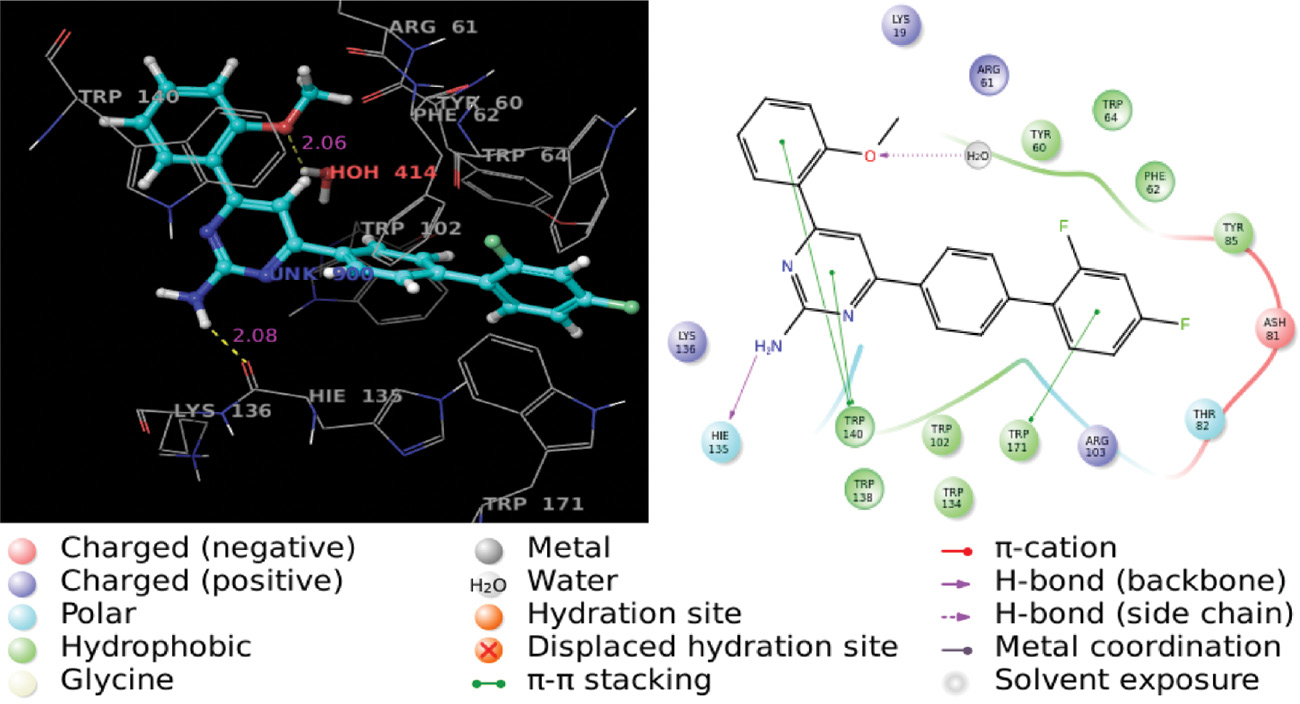
3D and 2D images of the complex of compound 5g with 4LRH protein residues.
Conclusions
Compounds 5a–g were synthesized and assessed for in vitro antimicrobial and cytotoxic activities, with the fluorine-substituted compound 5b having the highest activity. The in silico docking studies revealed that compounds 5a–g can inhibit the protein 4LRH and thus have great potential for development as anticancer agents.
Experimental
All reagents and solvents were of analytical grade and used without further purification. Melting points were measured in open glass capillaries and were uncorrected. The FT-IR spectra were recorded on an AVATAR-330 FT-IR spectrophotometer using KBr pellets. The NMR spectra were recorded in CDCl3 on a BRUKER Avance III 400 spectrometer operating at 400 MHz for 1H and 100 MHz for 13C. Mass spectra were recorded on a SCIEX-API 2000 spectrometer. Elemental analysis was carried out using a VARIOMICRO V2.2.0 CNH analyzer.
Antimicrobial assays
The clinical isolates of bacterial strains Staphylococcus aureus, Bacillus subtilis, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa, and a fungal strain Candida albicans were obtained from the Department of Microbiology, Rajah Muthiah Medical College and Hospital, Annamalai University, Annamalainagar, Tamilnadu, India. The strains were inoculated on a sterile medium and sub-cultured on Mueller Hinton agar plates and maintained on agar at 4°C. Gentamycin was used as a positive control for bacteria and fungi.
Disc diffusion assay
Antimicrobial activity was assayed using the standard disc diffusion method. Inhibition zones were measured and compared with the standard positive controls. All the tests were carried out in triplicate.
Minimum inhibitory concentration (MIC)
The dilution susceptibility testing method was used for MIC determination with reference to the literature [20]. The test compounds were dissolved in 1 mL of chloroform. The growth was indicated by the measure of a white pellet on a well bottom.
Cell viability assay
The cytotoxicity of the synthesized compounds was tested against Hep-2 cell lines using the MTT assay [21]. The MTT assays were performed four times independently, and each independent experiment was done in triplicate. The percentage of survival was calculated using the formula:
Molecular docking
Molecular docking was performed with the aid of Maestro v. 9.3.5 of the Schrödinger software suite, 2011. The 3D crystallographic structures of proteins (PDB ID: 4LRH, 3ACX) were retrieved from Protein Data Bank (www.rcsb.org/pdb). The protein structures were pre-processed and refined by Protein Preparation Wizard. Further, they were minimized by Optimized Potential for Liquid Simulations (OPLS)-2005 force field until the root mean square deviation (RMSD) reached the value of 0.3 Å. The ligands were optimized by a LigPrep program using the OPLS-2005 force field to generate the lowest energy state of ligands. The molecular docking studies of the ligand and protein were carried out by GLIDE. The best fit ligands with the target protein were ranked based on G score.
Synthesis of 4-(2′,4′-difluorobiphenyl-4-yl)-6-arylpyrimidin-2-amines 5a–g
A solution of (2E)-1-(2′,4’-difluorobiphenyl-4-yl)-3-arylprop-2-en-1-one (3a–g, 0.01 mol) and guanidine nitrate (4, 0.01 mol) in ethanol (30 mL) was treated with aqueous sodium hydroxide (10%, 2 mL) and the mixture was heated under reflux for 2 h. An additional amount of the sodium hydroxide solution was added and the mixture was heated for an additional 6 h. Progress of the reaction was monitored by thin layer chromatography (TLC). After cooling, the mixture was poured into crushed ice and the resultant solid was filtered out, washed with water, dried, and crystallized from ethanol.
4-(2′,4′-Difluorobiphenyl-4-yl)-6-phenylpyrimidin-2-amine (5a)
Yellow solid; yield 85%; mp 104–106°C; IR: 3495 (N-H), 1631 (C=N), 2845–3047 (C-H), 1103, 1141 cm−1 (C-F); 1H NMR: δ 6.78 (s, 2H, NH2), 7.77 (s, 1H, H5), 7.20–8.34 (m, 12H, Ar-H); 13C NMR: δ 101.9 (C5), 104.3–137.3 (Ar-C), 159.2 (C4′), 162.1(C2′), 163.9 (C2), 164.2 (C4), 164.9 (C6); ESI-MS: m/z 360.5 [M+1]+. Anal. Calcd for C22H15F2N3: C, 73.53; H, 4.21; N, 11.69. Found: C, 73.34; H, 4.45; N, 11.38.
4-(2′,4’-Difluorobiphenyl-4-yl)-6-(4-fluorophenyl)pyrimidin-2-amine (5b)
Pale yellow solid; yield 87%; mp 144–146°C; IR: 3428 (N-H), 1639 (C=N), 2849–2958 (C-H), 1098, 1137 (C-F); 1H NMR: δ 6.80 (s, 2H, NH2), 7.78 (s, 1H, H5), 7.04–8.34 (m, 11H, Ar-H); 13C NMR: δ 101.6 C5, 104.4–136.7 (Ar-C), 159.1 (C4′), 161.5 (C2′), 163.8 (C2), 163.9 (C4), 164.3 (C6); ESI-MS: m/z 378.2 [M+1]+. Anal. Calcd for C22H14F3N3: C, 70.02; H, 3.74; N, 11.14. Found: C, 70.35; H, 3.98; N, 11.07.
4-(2′,4’-Difluorobiphenyl-4-yl)-6-(4-bromophenyl)pyrimidin-2-amine (5c)
Yellow solid; yield 79%; mp 96–98°C; IR: 3423 (N-H), 1637 (C=N), 2849–2958 (C-H), 1101, 1140 cm−1 (C-F); 1H NMR: δ 6.84 (s, 2H, NH2), 7.80 (s, 1H, H5), 7.22–8.34 (m, 11H, Ar-H); 13C NMR: δ 101.8 (C5), 104.3–140.6 (Ar-C), 159.1 (C4’), 161.9 (C2′), 163.8 (C2), 163.9 (C4), 164.5 (C6); ESI-MS: m/z 438.0 [M+1]+. Anal. Calcd for C22H14F2N3Br: C, 60.29; H, 3.22; N, 9.59. Found: C, 60.26; H, 3.59; N, 9.31.
4-(2′,4’-Difluorobiphenyl-4-yl)-6-(4-chlorophenyl)pyrimidin-2-amine (5d)
Yellow solid; yield 82%; mp 128–130 °C; IR: 3493 (N-H), 1636 (C=N), 2855–3080 (C-H), 1101, 1141 (C-F) cm−1; 1H NMR: δ 6.75 (s, 2H, NH2), 7.78 (s, 1H, H5), 7.22–8.33 (m, 11H, Ar-H); 13C NMR: δ 101.8 (C5), 104.4–140.2 (Ar-C), 159.1 (C4′), 161.9 (C2′), 163.7 (C2), 163.9 (C4), 164.5 (C6); MS: m/z 394.2 [M+1]+. Anal. Calcd for C22H14F2N3Cl: C, 67.10; H, 3.58; N, 10.67. Found: C, 67.03; H, 3.83; N, 10.42.
4-(2′,4′-Difluorobiphenyl-4-yl)-6-(4-methylphenyl)pyrimidin-2-amine (5e)
Pale brown solid; yield 76%; mp 136–138°C; IR: 3482 (N-H), 1628 (C=N), 2918–3041(C-H), 1104, 1142 cm−1 (C-F); 1H NMR: δ 2.38 (s, 3H, CH3), 6.74 (s, 2H, NH2), 7.73 (s, 1H, H5), 7.0–8.32 (m, 11H, Ar-H); 13C NMR: δ 20.9 (CH3), 101.5 C5, 104.4–140.3 (Ar-C), 159.1 (C4′), 162.1 (C2′), 163.9 (C2), 164.1 (C4), 164.9 (C6); ESI-MS: m/z 374.1 [M+1]+. Anal. Calcd for C23H17F2N3: C, 73.98; H, 4.59; N, 11.25. Found: C, 73.87; H, 4.77; N, 10.93.
4-(2′,4’-Difluorobiphenyl-4-yl)-6-(3-nitrophenyl)pyrimidin-2-amine (5f)
Brown solid; yield 89%; mp 162–164°C; IR: 3491 (N-H), 1637 (C=N), 2851–3082 (C-H), 1101, 1141 cm−1 (C-F); 1H NMR: δ 6.97 (s, 2H, NH2), 7.96 (s, 1H, H5), 7.22–9.07 (m, 11H, Ar-H); 13C NMR: δ 102.2 C(5), 104.3–148.3 (Ar-C), 159.1 C(4’), 161.7 C(2′), 163.1 C(2), 163.9 C(4), 164.9 C(6); ESI-MS: m/z: 405.4 [M+1]+. Anal. Calcd for C22H14F2N4O2: C, 65.35; H, 3.49; N, 13.86. Found: C, 64.97; H, 3.71; N, 13.98.
4-(2′,4’-Difluorobiphenyl-4-yl)-6-(2-methoxyphenyl)pyrimidin-2-amine (5g)
Yellow solid; yield 78%; mp 130–132°C; IR: 3450 (N-H), 1597 (C=N), 2837–3072 (C-H), 1100, 1138 cm−1 (C-F); 1H NMR: δ 3.85 (s, 3H, OCH3), 6.69 (s, 2H, NH2), 7.79 (s, 1H, H5), 7.01–8.06 (m, 11H, Ar-H); 13C NMR: δ 55.4 (OCH3), 96.9 C(5), 104.1–138.8 (Ar-C), 159.1 C(4′), 161.6 C(2′), 163.0 C(2), 163.9 C(4), 164.5 C(6); ESI-MS: m/z 392.3 [M+3]+. Anal. Calcd for C23H17F2N3O: C, 70.94; H, 4.40; N, 10.79. Found: C, 70.58; H, 4.69; N, 10.50.
Supplementary information
Tabulated biological activities of all compounds, 2-D NMR of compound 5a, and docking poses of all compounds are given in the online Supplement.
Acknowledgments
We express our sincere thanks to the Department of Chemistry, Annamalai University, Annamalainagar, Tamilnadu, India, for providing laboratory and library facilities.
References
[1] Dudhe, R.; Sharma, P. K.; Verma, P.; Chaudhary, A. Pyrimidine as anticancer agent: a Review. J. Adv. Sci. Res.2011, 2, 10–17.10.4103/0976-9234.90213Search in Google Scholar
[2] Baker, S. J.; Premkumar Reddy, E. Targeted inhibition of kinases in cancer therapy. Mt Sinai. J. Med. 2010, 77, 573–586.10.1002/msj.20220Search in Google Scholar
[3] Koroleva, E. V.; Gusak, K. N.; Ignatovich, Z. V. Synthesis and applications of 2-aminopyrimidine derivatives as key intermediates in chemical synthesis of biomolecules. Russ. Chem. Rev. 2010, 79, 655–681.10.1070/RC2010v079n08ABEH004116Search in Google Scholar
[4] Nagaraj, A.; Reddy, S. C. Synthesis and biological study of novel bis-chalcones, bis-thiazines and bis-pyrimidines. J. Iran. Chem. Soc. 2008, 5, 262–267.10.1007/BF03246116Search in Google Scholar
[5] Atwal, K. S.; Swanson, B. N.; Moreland, S. Dihydropyrimidine calcium channel blockers. 3, 3-Carbamoyl-4-aryl-1,2,3,4-tetrahydro-6-methyl-5-pyrimidinecarboxylic acid esters as orally effective antihypertensive agents. J. Med. Chem. 1991, 34, 806–811.10.1021/jm00106a048Search in Google Scholar
[6] Nimavat, K. S.; Popat, K. H.; Vasoya, S. L.; Joshi, H. S. Synthesis, anticancer, antitubercular and antimicrobial activity of 1-substituted 3-aryl-5-(3′-bromophenyl)-pyrazolines. Indian. J. Heterocycl. Chem. 2003, 12, 225–228.Search in Google Scholar
[7] Vashi, K.; Naik, H. B. Synthesis and antibacterial activity of some novel chalcones and pyrimidine-2-one derivatives. Asian J. Chem. 2005, 17, 240–244.Search in Google Scholar
[8] Amir, M.; Javed, S. A.; Kumar, H. Synthesis and biological evaluation of some 4-(1H-indol-3-yl)-6-phenyl-1,2,3,4-tetrahydropyrimidin-2-ones/thiones as potent anti-inflammatory agents. Acta Pharm. 2008, 58, 467–477.10.2478/v10007-008-0028-xSearch in Google Scholar
[9] Ziessel, R.; Lehn, J. M. Synthesis and metal-binding properties of polybipyridine ligands derived from acyclic and macrocyclic polyamines. Helv. Chim. Acta1990, 73, 1149–1162.10.1002/hlca.19900730502Search in Google Scholar
[10] Nimmanapalli, R.; Bryan, E. O.; Huang, M.; Bali, P.; Burnette, P. K.; Loughran, T.; Tepperberg, J.; Jove, R.; Bhalla, K. Molecular characterization and sensitivity of STI-571 (imatinib mesylate, Gleevec)-resistant, Bcr-Abl-positive, human acute leukemia cells to SRC kinase inhibitor PD180970 and 17-allylamino-17-demethoxygeldanamycin. Cancer Res.2002, 62, 5761–5769.Search in Google Scholar
[11] Bonacorso, H. G.; Wastowski, A. D.; Zanatta, N.; Martins, M. A. P.; Naue, J. A. Haloacetylated enol ethers 10. Condensation of β-alkoxyvinyl trifuoromethyl ketones with thiosemicarbazide. Synthesis of new trifuoromethyl 4,5-dihydro-1H-1-pyrazolethiocarboxyamides. J. Fluorine Chem.1998, 92, 23–26.10.1016/S0022-1139(98)00242-5Search in Google Scholar
[12] Deng, H.; Hagan, D. O.; Schaffrath, C. Fluorometabolite biosynthesis and the fluorinase from Streptomyces cattleya. Nat. Prod. Rep. 2004, 21, 773–784.10.1039/b415087mSearch in Google Scholar PubMed
[13] Isanbor, C.; Hagan, D. O. Fluorine in medicinal chemistry: A review of anti-cancer agents. J. Fluorine Chem.2006, 127, 303–319.10.1016/j.jfluchem.2006.01.011Search in Google Scholar
[14] Kirk, K. L. Fluorine in medicinal chemistry: Recent therapeutic applications of fluorinated small molecules. J. Fluorine Chem.2006, 127, 1013–1029.10.1016/j.jfluchem.2006.06.007Search in Google Scholar
[15] Thanusu, J.; Kanagarajan, V.; Gopalakrishnan, M. 4-(4-Morpholinophenyl)-6-arylpyrimidin-2-amines: synthesis, spectral analysis, and in vitro microbiological evaluation. J. Enzyme Inhib. Med. Chem. 2010, 25, 347–353.10.3109/14756360903179468Search in Google Scholar
[16] Balasankar, T.; Nagarajan, S. Synthesis and antibacterial activities of some 2-amino-4,6- diarylpyrimidines. Heterocycl. Commun. 2004, 10, 451–456.10.1515/HC.2004.10.6.451Search in Google Scholar
[17] Fathimunnisa, M.; Manikandan, H.; Selvanayagam, S. Synthesis of novel (2E)-1-[4-(2,4-difluorophenyl)phenyl]3-arylprop-2-en-1-ones: Investigation on spectral, antibacterial, molecular docking and theoretical studies. J. Mol. Struct. 2015, 1099, 407–418.10.1016/j.molstruc.2015.06.078Search in Google Scholar
[18] Fargualy, A. M.; Habib, N. S.; Ismail, K. A.; Hassan, A. M.; Sarg, M. T. Synthesis, biological evaluation and molecular docking studies of some pyrimidine derivatives. Eur. J. Med. Chem. 2013, 66, 276–295.10.1016/j.ejmech.2013.05.028Search in Google Scholar
[19] Shah, A.; Khan, A. M.; Qureshi, R.; Ansari, F. L.; Nazar, M. F.; Shah, S. S. Redox Behavior of anticancer chalcone on a glassy carbon electrode and evaluation of its interaction parameters with DNA. Int. J. Mol. Sci. 2008, 9, 1424–1434.10.3390/ijms9081424Search in Google Scholar
[20] Andrews, J. M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16.10.1093/jac/48.suppl_1.5Search in Google Scholar
[21] Denizot, S.; Lang, R. Rapid colorimetric assay for cell growth and survival. J. Immunol. Methods1986, 89, 271–277.10.1016/0022-1759(86)90368-6Search in Google Scholar
Supplemental Material:
The online version of this article (DOI: 10.1515/hc-2016-0050) offers supplementary material, available to authorized users.
©2016 Walter de Gruyter GmbH, Berlin/Boston
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Articles in the same Issue
- Frontmatter
- Research Articles
- Pyrimidinethione as a building block in heterocyclic synthesis: synthesis of pyrano[2,3-d]pyrimidine, chromeno[2,3-d]pyrimidine, pyrido[3′,2′:5,6]pyrano[2,3-b]pyridine, and pyrimido[5′,4′:5,6]pyrano[2,3-d]pyrimidine derivatives
- C1-Substituted N-tert-butoxycarbonyl-5-syn-tert-butyldimethylsilyloxymethyl-2-azabicyclo[2.1.1]hexanes as conformationally constrained β-amino acid precursors
- Synthesis and characterization of 1,3,4-thiadiazole-2,5-dithio crown ethers
- Oxidative reaction of 2-aminopyridine-3-sulfonyl chlorides with tertiary amines
- New 8-substituted BODIPY-based chromophores: synthesis, optical and electrochemical properties
- Heterocyclization of 5,6-disubstituted 3-alkenyl-2-thioxothieno[2,3-d]pyrimidin-4-one with p-alkoxyphenyltellurium trichloride
- Synthesis and biological evaluation of 4-(2′,4′-difluorobiphenyl-4-yl)-6-arylpyrimidin- 2-amine derivatives
- Synthesis and antimicrobial properties of cycloheptyl substituted benzimidazolium salts and their silver(I) carbene complexes
- Solvent-free microwave-assisted synthesis and biological evaluation of 2,2-dimethylchroman-4-one based benzofurans
Articles in the same Issue
- Frontmatter
- Research Articles
- Pyrimidinethione as a building block in heterocyclic synthesis: synthesis of pyrano[2,3-d]pyrimidine, chromeno[2,3-d]pyrimidine, pyrido[3′,2′:5,6]pyrano[2,3-b]pyridine, and pyrimido[5′,4′:5,6]pyrano[2,3-d]pyrimidine derivatives
- C1-Substituted N-tert-butoxycarbonyl-5-syn-tert-butyldimethylsilyloxymethyl-2-azabicyclo[2.1.1]hexanes as conformationally constrained β-amino acid precursors
- Synthesis and characterization of 1,3,4-thiadiazole-2,5-dithio crown ethers
- Oxidative reaction of 2-aminopyridine-3-sulfonyl chlorides with tertiary amines
- New 8-substituted BODIPY-based chromophores: synthesis, optical and electrochemical properties
- Heterocyclization of 5,6-disubstituted 3-alkenyl-2-thioxothieno[2,3-d]pyrimidin-4-one with p-alkoxyphenyltellurium trichloride
- Synthesis and biological evaluation of 4-(2′,4′-difluorobiphenyl-4-yl)-6-arylpyrimidin- 2-amine derivatives
- Synthesis and antimicrobial properties of cycloheptyl substituted benzimidazolium salts and their silver(I) carbene complexes
- Solvent-free microwave-assisted synthesis and biological evaluation of 2,2-dimethylchroman-4-one based benzofurans

