CYP2D6 and CYP3A4 variants influence the risk and outcome of COVID-19 infection among rheumatoid arthritis patients maintained on hydroxychloroquine
-
Mohammad Salem Hareedy
, Sonya Mohamed Rashad
Abstract
Objectives
Hydroxychloroquine (HCQ) has been used as an off label for the management of coronavirus disease (Covid-19) infection with other drugs. However, different genetic variants can affect the metabolism of HCQ leading to inter-individual differences in its efficacy. In this study, we investigated the effects of variants in CYP2D6, CYP3A4 and CYP3A5 on the risk of Covid-19 infection among patients receiving HCQ for controlling rheumatoid arthritis (RA).
Methods
A total of 60 patients were genotyped for CYP2D6*2XN, CYP2D6*4, CYP3A4*1B and CYP3A5*2. They were receiving HCQ for the treatment of RA. The patients were evaluated clinically for fever and dry cough, radiologically via chest computed tomography (CT) and immunologically via anti-Covid-19 IgG and IgM titers.
Results
Variants in CYP2D6 significantly affected the grade of ground glass (CYP2D6*4 AA carriers showed the higher risk for grade 3) and the risk of positive anti-Covid-19 IgM (CYP2D6*2XN CC and CYP3A4*1B AA had the lowest risk), the duration of HCQ, the use of corticosteroids or gender did not affect the Covid-19 status significantly.
Conclusions
In general, the outcome of the studied patients receiving HCQ was good (no deaths, no intubation needed). CYP2D6 variants could affect the outcome of Covid-19 infection.
Introduction
A great debate about the role of the disease modifying anti rheumatic (DMAR) agents chloroquine and hydroxychloroquine (HCQ) was raised after the coronavirus disease 19 (Covid-19) pandemic [1]. HCQ is used for the treatment of malaria, rheumatoid arthritis (RA), systemic lupus erythematous (SLE) and to prevent thrombosis among patients with anti-phospholipid antibody syndrome [2], [3].
HCQ has more safety profile and is more commercially and pharmaceutically available than Chloroquine [4]. Both the drugs were used to control symptomatic patients suffering from Covid-19 with other drugs like azithromycin and different antiviral agents [5]. In spite of the scientific efforts; still there is no definitive approved drug or vaccine for treatment or prevention of Covid-19 [6].
Chloroquine and HCQ got a temporary permission under the title “emergency use authorization” for use in the treatment of Covid-19 in hospitalized patients (not for prophylaxis) in March 2020 from the United States Food and Drug Administration (FDA) [7], and then this permission was revoked by the FDA in the mid-June 2020 due to lack of evidences that HCQ can affect Covid-19 course (WWW.fda.gov). However, patients already receiving HCQ for management of RA are good candidates to evaluate the potential beneficial effects of HCQ during Covid-19 pandemic.
Patients with RA receiving immune suppressive drugs (especially corticosteroids) or DMAR drugs could be at higher risk for infections in general and Covid-19 during the pandemic. However, the risk or the incidence of Covid-19 infections among these patients couldn’t be easily concluded [8], [9].
Patients receiving steroids could be at a greater risk to get infections (bacterial, fungal, and viral) due to immunosuppression. On the other hand, corticosteroids could suppress inflammatory responses. The exact relation between steroids and risk or Covid-19 infection and its course has still not been understood [8].
The use of analgesic antipyretics (like acetaminophen) or nonsteroidal anti-inflammatory drugs for the treatment of pain and arthritis in patients with RA may obscure fever and delay the diagnosis of Covid-19. Ibuprofen use for treatment of active arthritis might worsen Covid-19 infection due to high uptake of viruses by pneumocytes [8]. So, patients receiving these drugs for the treatment of RA should be evaluated in the laboratory and radiologically to exclude Covid-19 infection.
An important factor which may explain the difficulty to conclude is the inter-individual variability in HCQ pharmacokinetics. The genetic variants in the metabolizing enzymes Cytochrome P450 2D6 (CYP2D6), 3A4 and 3A5 could alter the levels and ration of HCQ and its active metabolites and hence affect its total therapeutic efficacy [10], [11].
The questions raised now are: Does HCQ therapy for RA patients protect them from Covid-19 or even protect against Covid-19 pneumonia and respiratory complications? Why is the reported data not conclusive in such issue?
HCQ is extensively metabolized in the liver mainly via CYP2D6, 3A4, 3A5 and 2C8 [12], [13] The three major metabolites of HCQ are desethylchloroquine (DCQ), desethylhydroxychloroquine (DHCQ) and bisdesethylchloroquine. These metabolites were found to have similar activities to HCQ itself [14]. HCQ and its metabolites are extensively distributed in tissues including lungs, retina and skeletal muscles. The duration of HCQ therapy might have a role in its effectiveness [15].
The CYP2D gene is highly polymorphic among different populations [16]. Some of these variants were associated with decreased or even absent enzyme activity (e.g. CYP2D6*3,*4,*5, and *6) while some variants led to the production of more copies of CYP2D6 and hence increased metabolism (e.g. CYP2D6*2XN) [17], [18].
The aim of the present study was to evaluate the RA patients, who were maintained on HCQ, for Covid-19 infection through the clinical symptoms and signs, chest computed tomography (CT) and antibodies (IgM and IgG) related to Covid-19 pandemic and then to test these evaluations for a possible association with the CYP2D6*4, CYP2D6*2XN, CYP3A4*1B, CYP3A5*2, and CYP3A5*3 variants and finally to correlate the duration of HCQ therapy with the titers of IgM and IgG for Covid-19.
Materials and methods
Patients
A total of 60 patients receiving 400 mg of HCQ sulfate (plaquenil®) daily, for controlling RA, were included. They were evaluated monthly at the rheumatology department, Assiut University before Covid-19 pandemic. Inclusion criteria: Patients diagnosed previously to have RA and were taking HCQ regularly for the last six months. Exclusion criteria: patients who were incompliance with HCQ therapy.
During the Covid-19 pandemic (March–May 2020), all patients were evaluated (at a single visit) for fever, dry cough, and other symptoms suggestive for Covid-19 like malaise, fatigue, skin eruptions, and diarrhea. Blood samples were obtained for the routine blood picture, ALT, AST, albumin, ESR and for anti-Covid-19 IgG and IgM antibodies.
A CT for the chest for all patients was done to detect any changes linked to Covid-19 infection. Informed consents were obtained from the patients and the study was carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). The study is registered on ClinicalTrials.gov Identifier: NCT04389320 for Assiut University, Egypt.
The patients who were symptomatic for Covid-19 infection or had a positive anti-Covid-19 IgG or IgM were followed up via telephone calls (to decrease risk of spread of Covid-19) to evaluate their improvement.
Genotyping
DNA was extracted from EDTA-treated blood using the QIAmp® DNA Blood Mini Kit (Qiagen, USA). A Nanodrop spectrophotometer was used to determine the concentration and purity of the extracted DNA.
Two single nucleotide polymorphisms (SNPs) in the CYP2D6 gene were selected, the first SNP was the CYP2D6*4 (rs3892097; 1846G>A) and the second SNP was the CYP2D6*2XN (rs1135840; 4180G>C). These two SNPs were selected because they affect CYP2D6 extensively and in an opposite manner and because they were frequent among different populations including Caucasians and middle Eastern [18].
Three other SNPs— CYP3A4*1B (rs2740574; 392A>G), CYP3A5*3 (rs4362691; 6986A>G) and CYP3A5*3 (rs28365083; 27289C>A) were selected based on their frequencies and association with changes in CYP3A4 and CYP3A5 activity.
The genotypes were determined by TaqMan discrimination assays using the Type-it Fast Probe polymerase chain reaction (PCR) Master Mix (Qiagen) and the 7500 Fast Real Time PCR (Applied BiosystemsCA, USA). The SNP assays used were as follows: for CYP2D6*2XN (rs1135840; catalog number “C__27102414_10” and context sequence [VIC/FAM]: AGCACAAAGCTCATAGGGGGATGGG[C/G]TCACCAGGAAAGCAAAGACACCATG. The genotypes were identified as GG, GC, and CC. VIC and FAM are two different fluorescent reporter dyes.
For CYP2D6*4 (rs3892097; catalog number “C__27102431_D0” and Context sequence [VIC/FAM]: AGACCGTTGGGGCGAAAGGGGCGTC[C/T]TGGGGGTGGGAGATGCGGGTAAGGG. The genotypes were identified in reverse manner to be GG, GA, and AA.
For CYP3A4*1B (rs2740574; catalog number “C___1837671_50” and Context sequence [VIC/FAM]: TAAAATCTATTAAATCGCCTCTCTC[C/T]TGCCCTTGTCTCTATGGCTGTCCTC. The genotypes were identified in reverse manner to be GG, GA, and AA.
For CYP3A5*2 (rs28365083; catalog number “C__30633862_10” and Context sequence [VIC/FAM]: CTTTGGGTCATGGTGAAGAGCATAA[G/T]TTGGAATCACCACCATTGACCCTTT. The genotypes were identified in reverse manner to be CC, CA, and AA.
For CYP3A5*3 (rs776746; catalog number “C__26201809_30” and Context sequence [VIC/FAM]: ATGTGGTCCAAACAGGGAAGAGATA[T/C]TGAAAGACAAAAGAGCTCTTTAAAG.
The genotypes were identified in reverse manner to be GG, GA, and AA.
Measurement of Anti-Covid-19 IgG and IgM by ELISA
Serum levels of Anti-Covid-19 IgM and IgG were measured by ELISA Abnova (COVID-19 Human IgM IgG Assay Kit, Catalog Number KA5826 96 assays Version: 01) according to the manufacturer’s instructions.
CT of the chest
Parenchymal increased opacity was evaluated on the basis of its intensity and extent, parenchymal increased opacity was graded from 1 to 3 on the basis of the intensity [19]. Grade 1 corresponded to mild ground-glass opacity, and grade 2 corresponded to moderately increased opacity without obscuration of the vascular markings, while grade 3 corresponded to consolidation (i.e., marked increase in attenuation with obscuration of the vascular markings).
Li Guo et al. found that IgA and IgM antibodies were markers of acute Covid-19 infection (they appears within 3–6 days) while IgG appears later; after (10–18 days) of Covid-19 infections. They found measuring IgG Abs by ELISA as rapid, correlated to symptoms and can diagnose Covid-19 in cases of negative PCR or subclinical cases [20].
Our patients were classified according to the results of anti-covid-19 IgG and IgM into four categories or four Covid-19 statuses: those who have positive IgG and IgM antibodies were considered (had Covid-19 infection in the recent past), those who have negative for both IgG and IgM antibodies (did not catch Covid-19 infection), those who have positive IgG but negative IgM (passed cute Covid-19 infection) and those who have positive IgM but negative IgG (in the acute Covid-19 infection). All RA patients continued HCQ all over the period of the study.
Statistical analyses
The number and percentage of each studied genotype among each Covid-19 status were compared using X2 (Chi-square test). Each genotype was tested vs. the anti Covid-19 IgG, and the IgM results represented as “positive or negative” via Fisher exact test to compute relative risk in each.
The percentages of each studied genotype were tested vs. Covid-19 symptoms like fever and dry cough, vs. the need of hospitalization and vs. the degree or grade of ground glass in the chest CT using X2 (Chi-square test).
The quantitative titers of anti-Covid-19 IgM and IgG were tested vs. the studied genotypes of using one-way ANOVA and Bonferroni multiple comparison test. These titers were also correlated to the previous duration of HCQ therapy to test for protective cumulative effects of HCQ on risk of Covid-19 infection.
Finally, we tested the possible effects of previous corticosteroid therapy (for RA) on the Covid-19 statuses through comparing the percentages of each status among corticosteroid receivers and non-receivers and compare these data via X2 (Chi-square test). All statistical analyses were done using GraphPad Prism 7software (GraphPad, San Diego, CA).
Results
Patients demographics
Table 1 shows patients’ demographic characteristics and some laboratory investigations.
Patients’ demographics.
| Total number of patients maintained on hydroxychloroquine for management of rheumatoid arthritis n=60 |
| Age, years 48.45 ± 10.35 (20–68) |
| Gender Males (n=6; 10%) Females (n=54; 90%) |
| Body weight, kg 72.78 ± 9.913 (55–97) |
| Smoking history (past and present) Yes; n=20 No; n=40 |
| Duration of rheumatoid arthritis, years 11.35 ± 5.626 (2–25) |
| Duration of HQ treatment, years 7.833 ± 4.093 (2–23) |
| Receiving MTX Yes; n=46 No; n=14 |
| Receiving steroids (prednisone or prednisolone; 5–60 mg/day) Yes; n=37 No; n=23 |
| Hypertensive Yes; n=38 No; n=22 |
| Diabetes Yes; n=18 No; n=42 |
| Aspartate transaminase (AST), U/L 21.94 ± 15.55 (10–120) N.B. all patients have AST below 45 U/L except one patient (AST = 120 U/L) |
| Alanine transaminase (ALT), U/L 22.61 ± 13.46 (7–98) N.B. all patients have ALT below 56 U/L except one patient (ALT = 98 U/L) |
| Albumin, mg/dL 36.38 ± 5.491 (20–47) |
| WBCs, ×103/mm3 5.032 ± 2.215 (2.2–10.8) |
| Hemoglobin, g/dL 11.56 ± 1.656 (6.8–16.3) |
| Platelets, ×103/mm3 263.2 ± 87.85 (95–554) |
| Erythrocyte sedimentation rate first hour (ESR1), mm/h 38.87 ± 19.58 (12–103) |
Data are presented as mean ± standard deviation and a range, frequencies, or percentages.
Genotyping
The distribution of CYP2D6* 2XN genotypes among the studied patients were GG=26 (43.3%), GC=30 (50%) and CC=4 (6.7%) with a minimal allele frequency (MAF) of 0.32 and the distribution was not deviated from Hardy Weinberg equilibrium (HWE; p-value of Fisher exact test=0.23). However, the distribution of CYP2D6*4 was deviated from HWE (p-value of Fisher exact test=0.00002); genotypes were GG=40 (approximately, 66.7%), GA=10 (approximately, 16.7%) and AA=10 (approximately, 16.7%) with a MAF equals 0.25. CYP3A4*1B was distributed as follow; AA=34 (56.7%) AG=14 (23.3%) and AA=12 (20%); MAF=0.33 and HWE (p-value of Fisher exact test=0.0002). No variants were detected for the CYP3A5*2 (CC=60 (100%), CA=0 and AA=0); CYP3A5*3 was distributed as follow; GG=37 (61.67%), GA=16 (26.67%) and AA=7 (11.67%); MAF=0.25 and HWE (p-value of Fisher exact test=0.025)
Clinical symptoms of Covid-19 infection
The number of patients complained from fever were 27 (45%), complained from dry cough were 33 (55%), complained from fatigue were 30 (50%), and none complained from diarrhea or skin eruptions. These manifestations were reported by the patients to exist within few days before the clinical visit, the reporting physician observed elevated body temperature (more than 37.2 °C) during the visit of only nine patients (15%). Patients reported productive cough and known to be smokers were considered to have no recent dry cough.
Anti-Covid-19 IgM and IgM antibodies and course of Covid-19 among RA patients relieving HCQ
Fourteen patients out of 60 (i.e., 23.3%) were negative for both anti-Covid-19 IgG and IgM Abs (titer <25), the rest of the patients (n=46; 76.7%) were positive for either anti-Covid-19 IgG and/or IgM Abs (titer 25 or above). Seventeen patients (28.3% of total included patients) were asymptomatic but had elevated anti-Covid-19 IgG.
The patients were classified according the anti-Covid-19 IgM and IgM status into four groups: those having positive IgM and IgG titer reflecting that they had the infection in the recent past (n=37), those having negative IgM and IgG titers (i.e., didn’t catch Covid-19 infection, n=14), those having negative IgM but positive IgG titers (i.e., Passed acute infection, n=5) and finally those having positive IgM but negative IgG titers (i.e., they are in the acute infection, n=4).
The patients who needed hospitalization for management of Covid-19 infection were five (i.e., 8.3% of all RA studied patients and 10.9% of positive COVID-19 patients), one patient only (1.76% of all RA studied patients) needed intensive care unit (ICU); the patient admitted to the ICU has the genotype CYP2D6*2XN GC, CYP2D6*4 AA, CYP3A4*1B AG, and CYP3A5*3 AA and showed grade 3 ground glass on chest CT and elevated ALT and AST. The mean duration of stay at the hospital and ICU was 24.7 ± 4.1 days.
The nonhospitalized patients received treatment at home and no complications were reported during the 3–4 weeks from the clinical visit. All hospitalized patients did not need intubation or mechanical ventilation and they left the quarantine hospital and ICU within 3–4 weeks.
The correlation between anti-Covid-19 IgM and IgG titers and the duration of HCQ treatment is shown in Figure 1. There was a trend for IgM titers to decrease with longer durations of HCQ treatment, but the correlation was not significant (R2=0.12, p=0.395).
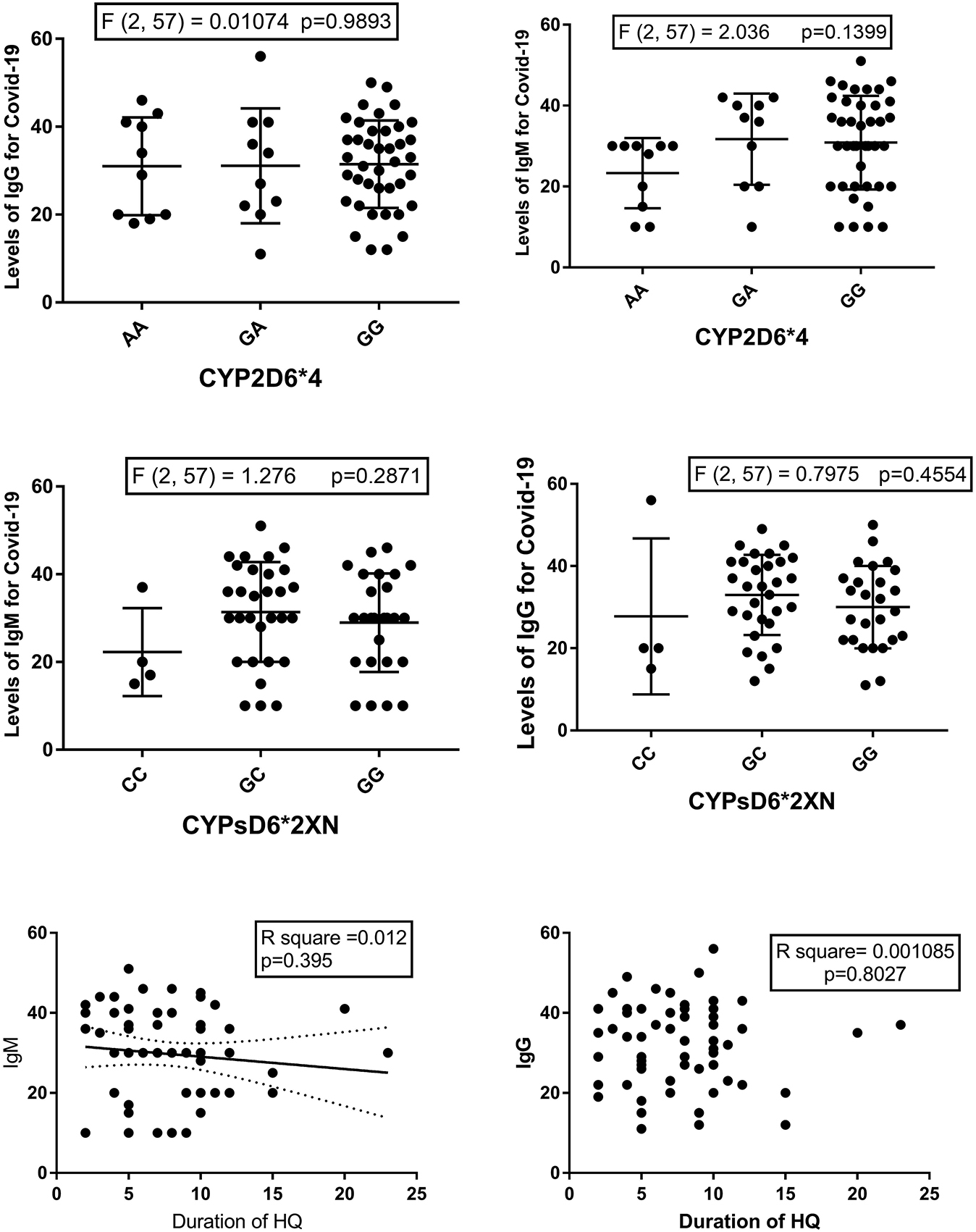
Levels of Anti-Covid-19 IgG and IgM vs. CYP2D6*4 (upper) and CYP2D6*2XN (middle) and the correlation between the duration of hydroxychloroquine (HCQ) treatment and levels of Anti-Covid-19 IgG and IgM (Lower).
The association between CYP2D6 genotypes and COVID-19 statuses
Table 2 shows the distribution of genotypes vs. anti-Covid-19 IgG and IgM status (positive or negative). Table 3 shows the results of Fisher exact test which compared each genotype with the other two genotypes for both anti-Covid-19 IgM and IgG status, where the Koopman asymptotic score was used to compute the relative risks. The CYP2D*2XN genotype CC carriers had the lower risk for a positive anti-Covid-19 IgG or IgM; however, the results were only significant when CC genotype compared to GC where, the latter was associated with 1.4 folds increased risk for positive anti-Covid-19 IgG (p=0.048).
Distribution of each CYP2D6*4, CYP2D6*2XN, CYP3A4*1B, and CYP3A5*3 genotypes in relation to Anti-Covid-19 IgG and IgM.
| SNP | Anti Covid-19 IgG | Anti Covid-19 IgM | ||
|---|---|---|---|---|
| Positive Total = 42 | Negative Total = 18 | Positive Total = 41 | Negative Total = 19 | |
| CYP2D6*4 number (percent to total number of patients) | AA = 6 (10%) | AA = 4 (6.7%) | AA = 6 (10%) | AA = 4 (6.7%) |
| AG = 6 (10%) | AG = 4 (6.7%) | AG = 7 (11.7%) | AG = 3 (5%) | |
| GG = 30 (50%) | GG = 10 (16.7%) | GG = 28 (46.7%) | GG = 12 (20%) | |
| CYP2D6*2XN number (percent to total number of patients) | GG = 17 (28.3%) | GG = 9 (15%) | GG = 18 (30%) | GG = 8 (13.3%) |
| GC = 24 (40%) | GC = 6 (10%) | GC = 22 (36.7%) | GC = 8 (13.3%) | |
| CC = 1 (1.7%) | CC = 3 (5%) | CC = 1 (1.7%) | CC = 3 (5%) | |
| CYP3A4*1B number (percent to total number of patients) | AA = 23 (38.3%) | AA = 11 (18.3%) | AA = 22 | AA = 12 (20%) |
| AG = 9 (15%) | AG = 3 (5%) | AG = 8 (13.3%) | AG = 4 (6.7%) | |
| GG = 10 (16.7%) | GG = 4 (6.7%) | GG = 11 (18.3%) | GG = 3 (5%) | |
| CYP3A5*3 number (percent to total number of patients) | GG = 25 (41.7%) | GG = 12 (20%) | GG = 25 (41.7%) | GG = 12 (20%) |
| GA = 12 (20%) | GA = 4 (6.7%) | GA = 11 (18.3%) | GA = 5 (8.3%) | |
| AA = 5 (8.3%) | AA = 2 (3.3%) | AA = 5 (8.3%) | AA = 2 (3.3%) | |
Positive means the titer is 25 and above. Negative means when the titer <25.
Relative risks (computed by Koopman asymptotic score) of CYP2D6, CYP3A4, and CYP3A5 genotypes in relation to anti-Covid-19 IgG and IgM results.
| Genotypes | Relative risk for positive IgM | 95% CI | p-Value (Fisher exact test) | Relative risk for positive IgG | 95% CI | p-Value (Fisher exact test) | |
|---|---|---|---|---|---|---|---|
| CYP2D6*2XN | GG vs. GC | 0.85 | 0.5016–1.574 | 0.7725 | 0.6911 | 0.411–1.27 | 0.2432 |
| GG vs. CC | 1.263 | 0.9408–2.04 | 0.2718 | 1.259 | 0.928–2.035 | 0.2742 | |
| GC vs. CC | 1.315 | 0.9973–2.213 | 0.0889 | 1.44 | 1.052–2.72 | 0.0480* | |
| CYP2D6*4 | AA vs. GG | 0.7059 | 0.2494–2.128 | 0.7067 | 0.5833 | 0.2094–1.752 | 0.4361 |
| AA vs. GA | 0.8077 | 0.3483–2.101 | >0.999 | 1 | 0.4222–2.62 | >0.999 | |
| GA vs. GG | 1 | 0.3346–3.297 | >0.999 | 0.5833 | 0.2094–1.752 | 0.4361 | |
| CYP3A4*1B | AA vs. AG | 0.7091 | 0.4475–1.014 | 0.0914 | 0.9148 | 0.6545–1.422 | 0.7294 |
| AA vs. GG | 0.5929 | 0.3687–0.8612 | 0.0103* | 0.9504 | 0.6651–1.509 | >0.9999 | |
| AG vs. GG | 0.7368 | 0.3429–1.883 | 0.6652 | 1.105 | 0.4791–3.24 | >0.9999 | |
| CYP3A5*3 | GG vs. GA | 0.9009 | 0.64–1.394 | 0.7484 | 0.9009 | 0.64–1.394 | 0.7484 |
| GG vs. AA | 0.9722 | 0.7463–1.416 | >0.9999 | 0.9722 | 0.7463–1.416 | >0.9999 | |
| GA vs. AA | 1.059 | 0.6128–2.449 | >0.9999 | 1.059 | 0.6128–2.449 | >0.9999 | |
Also, CYP3A4*1B AA genotype was significantly associated with decreased risk for a positive anti-Covid-19 IgM vs. GG genotypes but not vs. AG (Table 3).
Other genotypes did not show significant results (Table 3). However, the relative risks for a positive anti-Covid-19 IgM and IgG showed no difference between CYP2D6*4 GA carriers when compared to GG and AA carriers.
The titers of both IgG and IgM for COVID-19 were tested among different genotypes (Figures 1, 4, and 5). Results of either ordinary one-way ANOVA or Kruskal Wallis tests are shown in Figure 1. No significant associations were noticed.
There was no significant association between receiving steroids or not and the Covid-19 status among the studied patients, However, interestingly; higher percent of patients who passed Covid-19 infections and those who did not catch Covid-19 infection were among steroid receivers. There was no significant difference between male and female patients regarding the status of Covid-19 infection (X2, p=0.64).
The percentages of different Covid-19 statuses among different genotypes were plotted to show which of the genotypes has the highest or lowest existence among these status phenotypes. While the percentage of each genotype among different Covid-19 statuses were plotted to show the distribution of different Covid-19 statuses among each genotype (see Figures 2, 4, and 5).
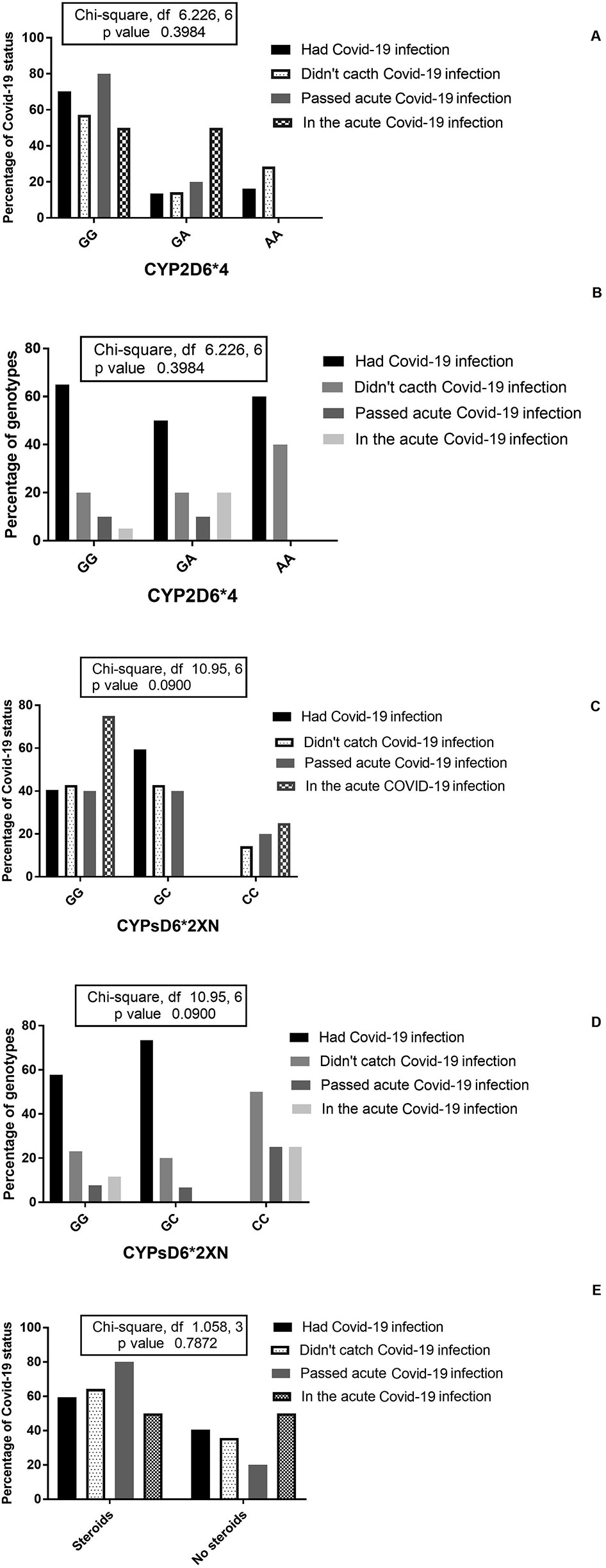
The percentage of Covid-19 statuses among CYP2D6*4 (A) and CYP2D6*2XN (C) genotypes and patients receiving steroids (E); the percentages of CYP2D6*4 (B) and CYP2D6*2XN (D) genotypes among different COVID-19 statuses.
Table 4 shows the distribution of genotypes vs. Covid-19 statuses (four categories). Figures 2, 4, and 5 show the results of X2 (Chi-Square test), where none of the carriers of the CYP2D6*2XN genotype GC fall in the status of the acute Covid-19 infection and 50% of carriers of the genotype CC didn’t catch Covid-19 infection, but p-value did not reach significant level (p=0.09). Carriers of AA genotype of the CYP2D6*4 showed higher percent of patients who did not catch the infection compared to GA carriers, but p-value was not significant (p=0.3984). Most of CYP3A5*3 GG genotypes did not catch Covid-19 infection and the percentage of patients who passed the acute Covid-19 infection was highest among CYP3A4*1B (AA genotype); but p-values were not significant (p<0.05).
Frequencies of patients in each genotype of the CYP2D6*2XN, CYP2D6*4, CYP3A4*1B and CYP3A5*3 in relation to symptoms (fever and dry cough), CT chest score for ground glass and the health care needed.
| Genotypes | CYP2D6*2XN | CYP2D6*4 | CYP3A4*1B | CYP3A5*3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GC | CC | AA | AG | GG | AA | AG | GG | GG | GA | AA | |
| Fever | 20.0% | 23.3% | 1.7% | 6.7% | 8.3 | 30.0% | 26.7% | 11.7% | 6.7% | 30.0% | 10.0% | 5.0% |
| No fever | 23.3% | 26.7% | 5.0% | 10.0% | 8.3% | 36.7% | 30.0% | 8.3% | 16.7% | 15.0% | 16.7 | 6.7% |
| Dry cough | 31.7% | 15.0% | 1.7% | 5.0% | 5.0% | 23.3% | 16.7% | 8.3% | 8.3% | 21.7% | 3.3 | 3.3% |
| No dry cough | 26.7% | 35.0% | 5.0% | 43.3% | 11.7% | 11.7% | 40.0% | 11.7% | 15.0% | 40.0% | 23.3 | 8.3% |
| ICU | 0.0% | 1.7% | 0.0% | 1.7% | 0.0% | 0.0% | 0.0% | 1.7% | 0.0% | 0.0% | 0.0% | 1.7% |
| Hospitalized | 5.0% | 1.7% | 1.7% | 1.7% | 3.3% | 3.3% | 5.0% | 1.7% | 3.3% | 6.7% | 1.7% | 1.7% |
| Non-hospitalized | 38.3% | 46.7% | 5.0% | 13.3% | 13.3% | 63.3% | 51.7% | 18.3% | 20.0% | 55.0% | 25.0% | 10.0% |
| CT score | ||||||||||||
| 0 | 28.3% | 31.7% | 5.0% | 11.7% | 11.7% | 41.7% | 38.3% | 11.7% | 15.0% | 36.7% | 20.0% | 6.7% |
| 1 | 11.7% | 16.7% | 0.0% | 3.3% | 1.7% | 23.3% | 16.7% | 6.7% | 5.0% | 20.0% | 5.0% | 3.3% |
| 2 | 3.3% | 0.0% | 1.7% | 0.0% | 3.3% | 1.7% | 1.7% | 0.0% | 3.3% | 3.3% | 1.7% | 8.3% |
| 3 | 0.0% | 1.7% | 0.0% | 1.7% | 0.0% | 0.0% | 0.0% | 1.7% | 0.0% | 0.0% | 0.0% | 1.7% |
ICU, Intensive care unit.
The ground glass score in chest CT was significantly associated with CYP2D6*4 genotype where the A (AA and GA) allele had higher percentages of grade 2 and 3 ground glass appearance than GG (p=0.048). The percentage of CYP2D6*2XN CC genotype was the highest among patients with normal chest CT (grade 0 ground glass) and at the same time in patients with grade 2 ground glass; but p-value was >0.5 (see Figure 3). There was no significant association between CT score and CYP3A4*1B and CYP3A5* three genotypes (Figures 4 and 5).
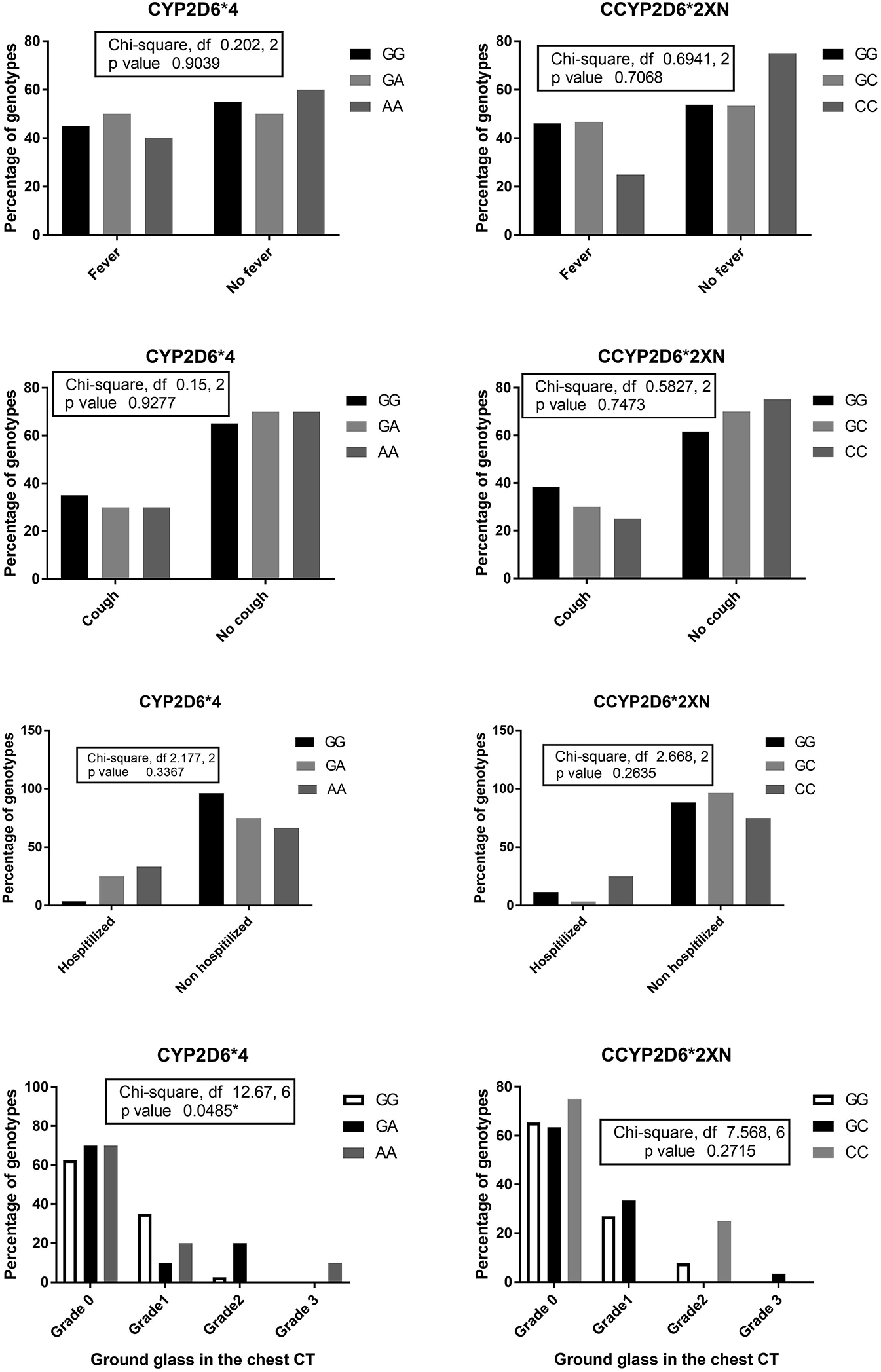
The percentages of CYP2D6*4 and CYP2D6*2XN genotypes among patients suffered from fever, dry cough, and those needed hospitalization and the percentages of different CYP2D6*4 and CYP2D6*2XN genotypes concerning the grade of ground glass in the chest CT.
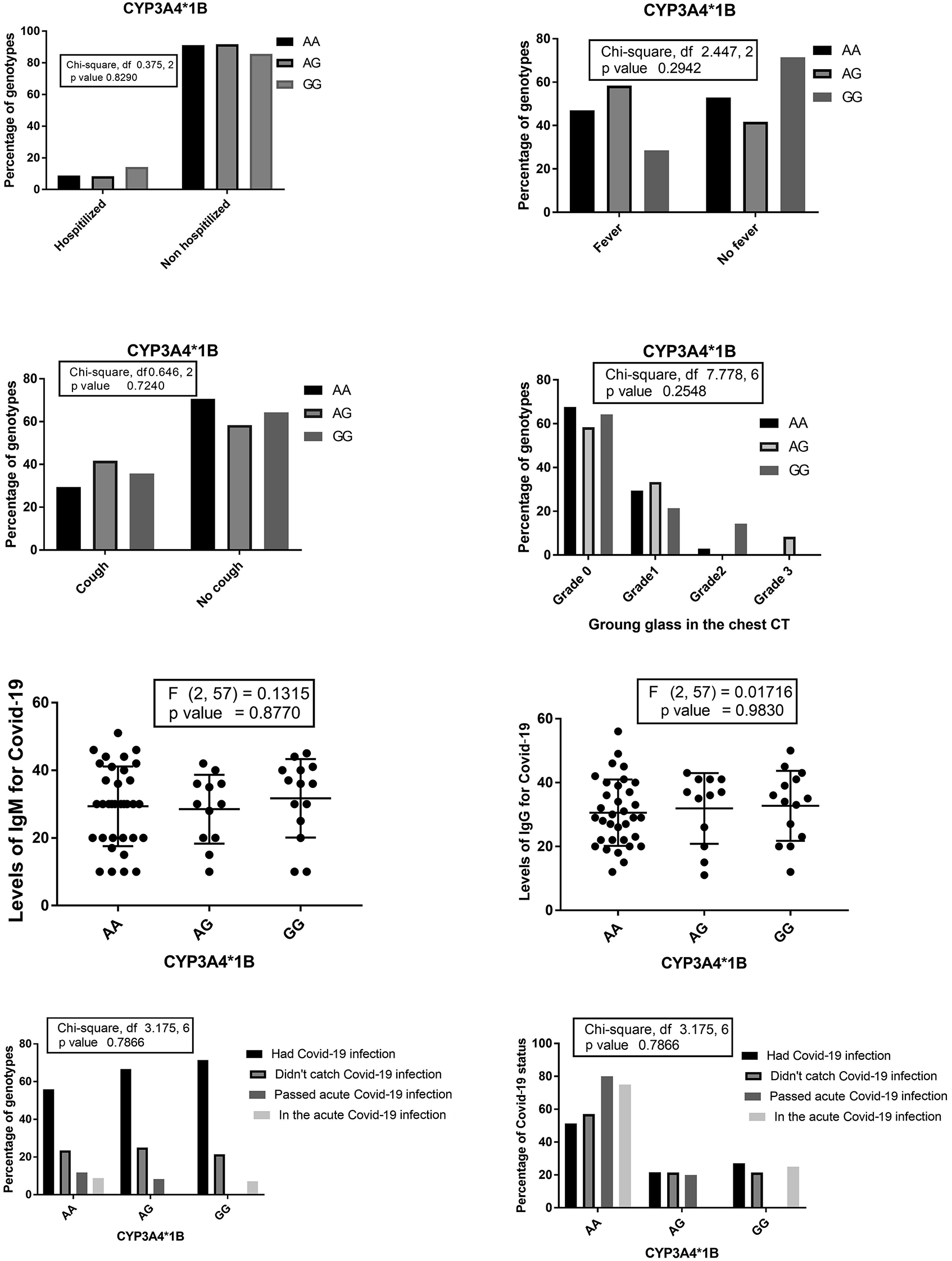
The percentages of different CYP3A4*1B genotypes among patients who needed hospitalization, suffered from fever, dry cough, and the percentages of CYP3A4*1B genotypes concerning the grade of ground glass in the chest CT. Levels of Anti-Covid-19 IgG and IgM among CYP3A4*1B genotypes and the percentage of different Covid-19 statuses among CYP3A4*1B genotypes the percentages of CYP3A4*1B genotypes among different Covid-19 statuses.
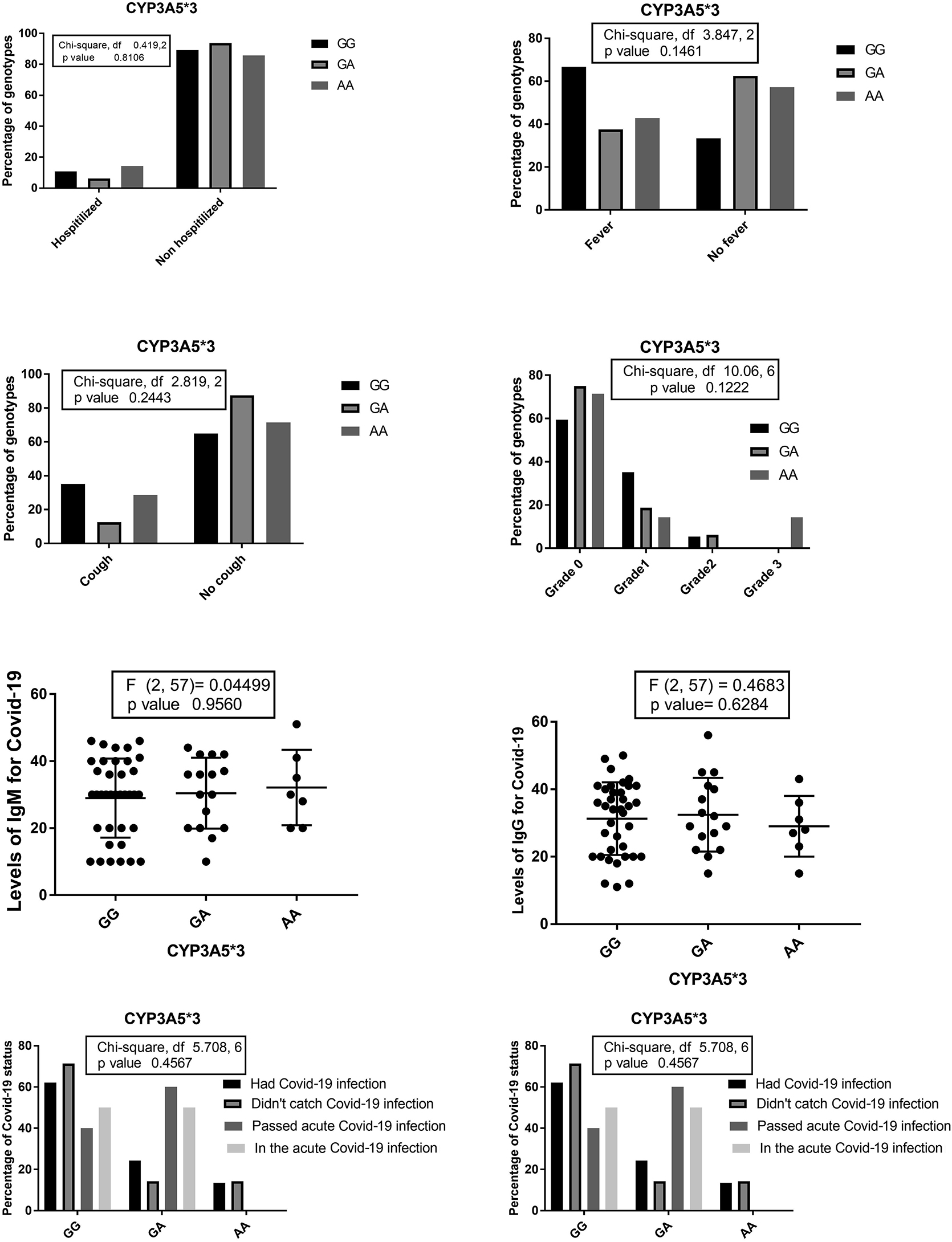
The percentages of different CYP3A5*3 genotypes among patients who needed hospitalization, suffered from fever, dry cough, and the percentages of CYP3A5*3 genotypes concerning the grade of ground glass in the chest CT. Levels of Anti-Covid-19 IgG and IgM among CYP3A5*3 genotypes and the percentage of different Covid-19 statuses among CYP3A5*3 genotypes the percentages of CYP3A5*3 genotypes among different Covid-19 statuses.
The percentage of patients who complained of fever, dry cough among different CYP2D6*4 did not differ obviously; these percentages were also statistically far from level of significance (see Figure 3). However, the percentages of hospitalization were higher among CYP2D6*4 (AA and GA) vs. GG, but statistically the significance of that difference couldn’t be confirmed (p>0.5; see Figure 3).
On the other hand, the percentage of patients who complained of fever and dry cough among CYP2D6*2XN genotypes were different obviously where the percent of CC genotype was the lowest among patients complained from fever and dry cough. At the same time, the percentage of CC genotype distribution was the highest among those patients who didn’t show fever or dry cough, but these percentages were statistically far from the level of significance (see Figure 3). However, the percentage of hospitalization was higher among CYP2D6*4 (AA and GA) vs. GG, but statistically the significance of that difference could not be confirmed (p>0.05) (Figure 3).
There was no statistical significant difference between the patients received who steroids and those did not receive and the status of Covid-19 infection (p=0.0787; Figure 2). However, and interestingly, the percentages of patients who did not catch Covid-19 (64%) and those who passed acute Covid-19 infection (80%) were higher among steroid receivers than those who didn’t receive steroid. At the same time, the percentages of patients who were in the acute Covid-19 infection were equal among steroid receivers than those who did not receive steroid (50% in each). The percentage of patients who had the Covid-19 infection in the recent past was higher among steroid receivers (59.5%).
Discussion
In the present study, the prevalence of Covid-19 infection among 60 RA patients receiving HCQ was about 76.7%. In contrast to the findings of Vastarella et al. who did not find symptoms suggestive of Covid-19 infection among 66 patients receiving HCQ for treatment of dermatological or rheumatologic diseases [21], Vastarella et al. pointed to possible protective effects of HCQ against Covid-19. But they did not test for Covid-19 infection (via anti-Covid IgG or IgM or PRC) since Covid-19 could be asymptomatic. The results of Vastarella et al. were not compared to control population not receiving HCQ.
The withdrawal of DMARDs except HCQ with addition of antiviral medications led to improvement of Covid-19 pneumonia in patients with RA [22]. HCQ was used for management of Covid-19 in many countries based on three major mechanisms; immunomodulation and effects on cytokines, prevention of minor thrombosis in the lungs and alteration of intracellular PH interfering with entry of virus particles [4], [23].
HCQ is safer than chloroquine and can suppress cytokine storm in patients with Covid-19 [24]. HCQ was found to be effective (and superior to chloroquine) in treatment of Covid-19 in the studies performed on cell cultures [25] and in clinical trials, HCQ lowered the viral loads in patients with Covid-19 infection [5], However, the role of HCQ for prevention of Covid-19 is not clear yet [26]. The active metabolites of HCQ contribute partially to its efficacy and side effects [15].
Induction of CYP2D6 is not documented and genetic variants in the CYP2D6 gene are the major cause of the large interindividual variation in CYP2D6 activity [27].
The CYP2D6 gene located on the long arm of the chromosome 22q13 is highly polymorphic [28], [29] and codes for the metabolizing enzyme CYP2D6 which participates greatly in the metabolism of both chloroquine and HCQ [12]. Carriers of CYP2D6*4 (1846G>A) have almost little to none CYP2D6 activity due to a splicing defect in an intron (between third and fourth exon) [30], [31], [32].
Our hypothesis is since, desethyl HCQ could have a greater role in the actions of HCQ. So, the rate of formation of desethyl HCQ (through CYP2D6 and other CYP families) could affect the covid-19 status among HCQ receivers for RA. CYP2D6*4 is the most common nonfunctional variant among CYP2D6 family. Previous studies linked an association between the CYP2D6*4 and treatment outcomes of several drugs like antiepileptic drugs, beta blockers, and antidepressant drugs [30], [31], [32].
The active metabolite desethyl HCQ was significantly associated with HCQ response in patients with RA [10]. Lee et al. found a significant association between certain CYP2D6*4 and *2XN variants and blood levels of HCQ and desethyl HCQ and their ratios in patients with SLE [30].
So, in the present study, CYP2D6*4 (AA) carriers were poor metabolizers and HCQ was metabolized poorly leading to decreased formation of one of the active metabolites (desethyl HCQ), this could decrease the efficiency of HCQ in preventing Covid-19 viruses from entering cells making the risk of Covid-19 infection and formation of IgM antibodies more likely.
Also, the levels of desethyl HCQ might be lower in carriers of CYP2D6*4 (AA) leading to a significant difference in the grade of ground glass opacities in the chest CT where CYP2D6*4 AA were the only patients who had grade 3 ground glass in the chest CT and the only admitted to ICU. CYP2D6*4 AA carriers also showed more hospitalization ratios vs. wild and heterozygous carriers (but p-value was >0.05).
CYP2D6*2XN was associated with increased enzyme activity through more expression of CYP2D6 copies leading to ultra-rapid metabolizer phenotype [18], [27]. Levels of desethyl HCQ are supposed to be higher among carriers of CYP2D6*2XN (Ultra-rapid metabolizers) and this could explain the lower risk for positive anti-Covid-19 IgM among these patients relative to GC genotype. Also, a greater percent of CYP2D6*2XN carriers didn’t experienced fever, dry cough or grade 3 ground glass in the chest CT (but p-values were >0.05).
CYP3A5*3 was not significantly associated with the risk or the course of covid-19 among the studied patients; however, CYP3A4*1B AA (normal enzyme activity and enhanced expression) genotype was associated with lower risk for anti-Covid 19 IgM levels vs. GG (associated with decreased activity in vivo as evaluated by Rosales et al. [33]). These findings also support our hypothesis.
Limitations and prospective
Limitations of the present study were the small number of included patients, the lack of information about glucose 6 phosphate dehydrogenase (G6PD) status among all included patients (however, no clinical or laboratory finding of hemolysis was detected in their available medical records). Investigating other CYP2D6 variants and CYP3A4 variants was another important limitation of the present study. Further pharmacogenomic evaluation will be interesting.
Conclusions
CYP2D6*2XN, CYP2D6*4, CYP3A4*1B, and CYP3A5*2 were analyzed in 60 patients receiving HCQ for management of RA. The patients were evaluated clinically, radiologically, and immunologically for the Covid-19. Some variants in CYP2D6 significantly affected the grade of ground glass (CYP2D6*4 AA carriers showed the higher risk for grade 3) and the risk of positive Anti-Covid-19 IgM (CYP2D6*2XN CC and CYP3A4*1B AA had the lowest risk), the duration of HCQ, the use of corticosteroids or gender didn’t affect the Covid-19 status significantly. In general, the outcome of the studied patients receiving HCQ was good (no deaths, no intubation needed). CYP2D6 variants are common among the studied patients in the present study and affected the outcome of covid-19 infection.
Research funding: None declared.
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
Competing interests: Authors state no conflict of interest.
Informed consent: Informed consent was obtained from all individuals included in this study.
Ethical approval: The study is registered on ClinicalTrials.gov Identifier: NCT04389320 for Assiut University, Egypt.
References
1. Ferner, RE, Aronson, JK. Chloroquine and hydroxychloroquine in covid-19. BMJ 2020;369:m1432. https://doi.org/10.1136/bmj.m1432.Suche in Google Scholar PubMed
2. Wang, TF, Lim, W. What is the role of hydroxychloroquine in reducing thrombotic risk in patients with antiphospholipid antibodies? Hematol Am Soc Hematol Educ Progr 2016;2016:714–6. https://doi.org/10.1182/asheducation-2016.1.714.Suche in Google Scholar PubMed PubMed Central
3. Alarcón, GS, McGwin, G, Bertoli, AM, Fessler, BJ, Calvo-Alén, J, Bastian, HM, LUMINA Study Group, et al.. Effect of hydroxychloroquine on the survival of patients with systemic lupus erythematosus: data from LUMINA, a multiethnic US cohort (LUMINA L). Ann Rheum Dis 2007;66:1168–72. https://doi.org/10.1136/ard.2006.068676.Suche in Google Scholar PubMed PubMed Central
4. Sahraei, Z, Shabani, M, Shokouhi, S, Saffaei, A. Aminoquinolines against coronavirus disease 2019 (COVID-19): chloroquine or hydroxychloroquine. Int J Antimicrob Agents 2020;55:105945. https://doi.org/10.1016/j.ijantimicag.2020.105945.Suche in Google Scholar PubMed PubMed Central
5. Gautret, P, Lagier, JC, Parola, P, Hoang, VT, Meddeb, L, Mailhe, M, et al.. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 2020;56:105949. https://doi.org/10.1016/j.ijantimicag.2020.105949.Suche in Google Scholar PubMed PubMed Central
6. Kupferschmidt, K, Cohen, J. Race to find COVID-19 treatments accelerates. Science 2020;367:1412–3. https://doi.org/10.1126/science.367.6485.1412.Suche in Google Scholar PubMed
7. Lenzer, J. Covid-19: US gives emergency approval to hydroxychloroquine despite lack of evidence. BMJ 2020;369:m1335. https://doi.org/10.1136/bmj.m1335.Suche in Google Scholar PubMed
8. Favalli, EG, Ingegnoli, F, De Lucia, O, Cincinelli, G, Cimaz, R, Caporali, R. COVID-19 infection and rheumatoid arthritis: faraway, so close!. Autoimmun Rev 2020;19:102523. https://doi.org/10.1016/j.autrev.2020.102523.Suche in Google Scholar PubMed PubMed Central
9. Monti, S, Balduzzi, S, Delvino, P, Bellis, E, Quadrelli, VS, Montecucco, C. Clinical course of COVID-19 in a series of patients with chronic arthritis treated with immunosuppressive targeted therapies. Ann Rheum Dis 2020;79:667–8. https://doi.org/10.1136/annrheumdis-2020-217424.Suche in Google Scholar PubMed PubMed Central
10. Munster, T, Gibbs, JP, Shen, D, Baethge, BA, Botstein, GR, Caldwell, J, et al.. Hydroxychloroquine concentration-response relationships in patients with rheumatoid arthritis. Arthritis Rheum 2002;46:1460–9. https://doi.org/10.1002/art.10307.Suche in Google Scholar PubMed
11. Wahie, S, Daly, AK, Cordell, HJ, Goodfield, MJ, Jones, SK, Lovell, CR, et al.. Clinical and pharmacogenetic influences on response to hydroxychloroquine in discoid lupus erythematosus: a retrospective cohort study. J Invest Dermatol 2011;131:1981–6. https://doi.org/10.1038/jid.2011.167.Suche in Google Scholar PubMed
12. Projean, D, Baune, B, Farinotti, R, Flinois, JP, Beaune, P, Taburet, AM, et al.. In vitro metabolism of chloroquine: identification of CYP2C8, CYP3A4, and CYP2D6 as the main isoforms catalyzing N-desethylchloroquine formation. Drug Metab Dispos 2003;31:748–54. https://doi.org/10.1124/dmd.31.6.748.Suche in Google Scholar PubMed
13. Kim, KA, Park, JY, Lee, JS, Lim, S. Cytochrome P450 2C8 and CYP3A4/5 are involved in chloroquine metabolism in human liver microsomes. Arch Pharm Res 2003;26:631–7. https://doi.org/10.1007/bf02976712.Suche in Google Scholar
14. Browning, DJ. Hydroxychloroquine and chloroquine retinopathy: screening for drug toxicity. Am J Ophthalmol 2002;133:649–56. https://doi.org/10.1016/s0002-9394(02)01392-2.Suche in Google Scholar PubMed
15. Chhonker, YS, Sleightholm, RL, Li, J, Oupický, D, Murry, DJ. Simultaneous quantitation of hydroxychloroquine and its metabolites in mouse blood and tissues using LC-ESI-MS/MS: an application for pharmacokinetic studies. J Chromatogr B Anal Tech Biomed Life Sci 2018;1072:320–7. https://doi.org/10.1016/j.jchromb.2017.11.026.Suche in Google Scholar PubMed PubMed Central
16. Ma, MK, Woo, MH, McLeod, HL. Genetic basis of drug metabolism. Am J Health Syst Pharm 2002;59:2061–9. https://doi.org/10.1093/ajhp/59.21.2061.Suche in Google Scholar PubMed
17. Evans, WE, Relling, MV. Moving towards individualized medicine with pharmacogenomics. Nature 2004;429:464–8. https://doi.org/10.1038/nature02626.Suche in Google Scholar PubMed
18. Qumsieh, RY, Ali, BR, Abdulrazzaq, YM, Osman, O, Akawi, NA, Bastaki, SM. Identification of new alleles and the determination of alleles and genotypes frequencies at the CYP2D6 gene in Emiratis. PloS One 2011;6:e28943. https://doi.org/10.1371/journal.pone.0028943.Suche in Google Scholar PubMed PubMed Central
19. Tokura, S, Akira, M, Okuma, T, Tazawa, R, Arai, T, Sugimoto, C, et al.. A semiquantitative computed tomographic grading system for evaluating therapeutic response in pulmonary alveolar proteinosis. Ann Am Thorac Soc 2017;14:1403–11. https://doi.org/10.1513/annalsats.201607-574oc.Suche in Google Scholar
20. Guo, L, Ren, L, Yang, S, Xiao, M, Chang, D, Yang, F, et al.. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 2020;71:778–85. 28. https://doi.org/10.1093/cid/ciaa310.Suche in Google Scholar PubMed PubMed Central
21. Vastarella, M, Patrì, A, Annunziata, MC, Cantelli, M, Nappa, P, Tasso, M, et al.. Can hydroxychloroquine be useful in the prevention of COVID-19? An Italian survey in dermatologic and rheumatologic patients already under treatment. J Am Acad Dermatol 2020;83:e77–9. https://doi.org/10.1016/j.jaad.2020.04.136.Suche in Google Scholar PubMed PubMed Central
22. Song, J, Kang, S, Choi, SW, Seo, KW, Lee, S, So, MW, et al.. Coronavirus Disease 19 (COVID-19) complicated with pneumonia in a patient with rheumatoid arthritis receiving conventional disease-modifying antirheumatic drugs. Rheumatol Int 2020;40:991–5. https://doi.org/10.1007/s00296-020-04584-7.Suche in Google Scholar PubMed PubMed Central
23. Ali, AS, Abdel-Rahman, MS, Almalikil, RS, Mohamed, AS, Alfaifi, KA, Fadil, AE, et al.. Optimizing the use of hydroxychloroquine in the management of COVID-19 given its pharmacological profile. J Pharmaceut Res Int 2020;32:29–43. https://doi.org/10.9734/jpri/2020/v32i830468.Suche in Google Scholar
24. Zhou, D, Dai, SM, Tong, Q. COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J Antimicrob Chemother 2020;75:1667–70. https://doi.org/10.1093/jac/dkaa114.Suche in Google Scholar PubMed PubMed Central
25. Yao, X, Ye, F, Zhang, M, Cui, C, Huang, B, Niu, P, et al.. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020;71:732–9. https://doi.org/10.1093/cid/ciaa237.Suche in Google Scholar PubMed PubMed Central
26. Cohen, MS. Hydroxychloroquine for the prevention of covid-19 - searching for evidence. N Engl J Med 2020;383:585–6. https://doi.org/10.1056/nejme2020388.Suche in Google Scholar PubMed PubMed Central
27. Ganoci, L, Božina, T, Mirošević Skvrce, N, Lovrić, M, Mas, P, Božina, N. Genetic polymorphisms of cytochrome P450 enzymes: CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5 in the Croatian population. Drug Metabol Personalized Ther 2017;32:11–21. https://doi.org/10.1515/dmpt-2016-0024.Suche in Google Scholar PubMed
28. Wilkinson, GR. Drug metabolism and variability among patients in drug response. N Engl J Med 2005;352:2211–21. https://doi.org/10.1056/nejmra032424.Suche in Google Scholar PubMed
29. Sistonen, J, Sajantila, A, Lao, O, Corander, J, Barbujani, G, Fuselli, S. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenetics Genom 2007;17:93–101. https://doi.org/10.1097/01.fpc.0000239974.69464.f2.Suche in Google Scholar PubMed
30. Lee, JY, Vinayagamoorthy, N, Han, K, Kwok, SK, Ju, JH, Park, KS, et al.. Association of polymorphisms of cytochrome P450 2D6 with blood hydroxychloroquine levels in patients with systemic lupus erythematosus. Arthritis Rheum 2016;68:184–90. https://doi.org/10.1002/art.39402.Suche in Google Scholar PubMed
31. Bijl, MJ, Visser, LE, van Schaik, RH, Kors, JA, Witteman, JC, Hofman, A, et al.. Genetic variation in the CYP2D6 gene is associated with a lower heart rate and blood pressure in beta-blocker users. Clin Pharmacol Ther 2009;85:45–50. https://doi.org/10.1038/clpt.2008.172.Suche in Google Scholar PubMed
32. Bijl, MJ, Visser, LE, Hofman, A, Vulto, AG, van Gelder, T, Stricker, BH, et al.. Influence of the CYP2D6*4 polymorphism on dose, switching and discontinuation of antidepressants. Br J Clin Pharmacol 2008;65:558–64. https://doi.org/10.1111/j.1365-2125.2007.03052.x.Suche in Google Scholar PubMed PubMed Central
33. Rosales, A, Alvear, M, Cuevas, A, Saavedra, N, Zambrano, T, Salazar, LA. Identification of pharmacogenetic predictors of lipid-lowering response to atorvastatin in Chilean subjects with hypercholesterolemia. Clin Chim Acta 2012;413:495–501. https://doi.org/10.1016/j.cca.2011.11.003.Suche in Google Scholar PubMed
© 2021 Walter de Gruyter GmbH, Berlin/Boston
Artikel in diesem Heft
- Frontmatter
- Reviews
- A review on the clinical trials of repurposing therapeutic drugs, mechanisms and preventive measures against SARS-CoV-2
- Drugs intervention study in COVID-19 management
- Original Articles
- CYP2D6 and CYP3A4 variants influence the risk and outcome of COVID-19 infection among rheumatoid arthritis patients maintained on hydroxychloroquine
- Metabolizer phenotype prediction in different Peruvian ethnic groups through CYP2C9 polymorphisms
- Pharmacogenetic algorithm for predicting daily dose of warfarin in Caucasian patients of Czech origin
- Fear of Hypoglycemia in Adults with diabetes mellitus switching to Treatment with IDegAsp Co-formulation to Examine real-world setting: an observational study (The HATICE study)
- Involvement of GABAergic and nitrergic systems in the anxiolytic and hypnotic effects of Curcuma longa: its interaction with anxiolytic-hypnotics
- Quality control, HPTLC analysis, antioxidant and antimicrobial activity of hydroalcoholic extract of roots of qust (Saussurea lappa, C.B Clarke)
- Recombinant human hyaluronidase PH20-mediated dermal spreading activity in mice is not altered by steroids, antihistamines, or salicylic acid
- Short Communication
- TCF7L2 rs7903146 C>T gene polymorphism is not associated with hypoglycemia in sulfonylurea-treated type 2 diabetic patients
- Case Report
- Spinal and cerebral hematoma in systemic lupus erythematosus and antiphospholipid syndrome: is drug interaction the culprit?
Artikel in diesem Heft
- Frontmatter
- Reviews
- A review on the clinical trials of repurposing therapeutic drugs, mechanisms and preventive measures against SARS-CoV-2
- Drugs intervention study in COVID-19 management
- Original Articles
- CYP2D6 and CYP3A4 variants influence the risk and outcome of COVID-19 infection among rheumatoid arthritis patients maintained on hydroxychloroquine
- Metabolizer phenotype prediction in different Peruvian ethnic groups through CYP2C9 polymorphisms
- Pharmacogenetic algorithm for predicting daily dose of warfarin in Caucasian patients of Czech origin
- Fear of Hypoglycemia in Adults with diabetes mellitus switching to Treatment with IDegAsp Co-formulation to Examine real-world setting: an observational study (The HATICE study)
- Involvement of GABAergic and nitrergic systems in the anxiolytic and hypnotic effects of Curcuma longa: its interaction with anxiolytic-hypnotics
- Quality control, HPTLC analysis, antioxidant and antimicrobial activity of hydroalcoholic extract of roots of qust (Saussurea lappa, C.B Clarke)
- Recombinant human hyaluronidase PH20-mediated dermal spreading activity in mice is not altered by steroids, antihistamines, or salicylic acid
- Short Communication
- TCF7L2 rs7903146 C>T gene polymorphism is not associated with hypoglycemia in sulfonylurea-treated type 2 diabetic patients
- Case Report
- Spinal and cerebral hematoma in systemic lupus erythematosus and antiphospholipid syndrome: is drug interaction the culprit?

