MedShapeNet – a large-scale dataset of 3D medical shapes for computer vision
-
Jianning Li
Abstract
Objectives
The shape is commonly used to describe the objects. State-of-the-art algorithms in medical imaging are predominantly diverging from computer vision, where voxel grids, meshes, point clouds, and implicit surface models are used. This is seen from the growing popularity of ShapeNet (51,300 models) and Princeton ModelNet (127,915 models). However, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D models of surgical instruments is missing.
Methods
We present MedShapeNet to translate data-driven vision algorithms to medical applications and to adapt state-of-the-art vision algorithms to medical problems. As a unique feature, we directly model the majority of shapes on the imaging data of real patients. We present use cases in classifying brain tumors, skull reconstructions, multi-class anatomy completion, education, and 3D printing.
Results
By now, MedShapeNet includes 23 datasets with more than 100,000 shapes that are paired with annotations (ground truth). Our data is freely accessible via a web interface and a Python application programming interface and can be used for discriminative, reconstructive, and variational benchmarks as well as various applications in virtual, augmented, or mixed reality, and 3D printing.
Conclusions
MedShapeNet contains medical shapes from anatomy and surgical instruments and will continue to collect data for benchmarks and applications. The project page is: https://medshapenet.ikim.nrw/.
Introduction
The success of deep learning in many fields of applications, including vision [1], language [2] and speech [3], is mainly due to the availability of large, high-quality datasets [4], [5], [6], such as ImageNet [7], CIFAR [8], Penn Treebank [9], WikiText [10] and LibriSpeech [11]. In 3D computer vision, Princeton ModelNet [12], ShapeNet [13], etc., are the de facto benchmarks for numerous fundamental vision problems, including 3D shape classification and retrieval [14], shape completion [15], shape reconstruction and segmentation [16]. Shape describes the geometries of 3D objects and is one of the most basic concepts in computer vision. Common 3D shape representations include point clouds, voxel occupancy grids, meshes, and implicit surface models (signed distance functions), which follow different data structures, cater for different algorithms, and are convertible to each other [17]. These shape representations diverge from gray-scale medical imaging data routinely used in clinical diagnosis and treatment procedures, such as computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), ultra sound (US), and X-ray.
The concept of shape in medical imaging is not novel. For example, statistical shape modeling (SSM) has been a longstanding method for medical image segmentation [18] and 3D anatomy modeling [19]. The use of shape priors and constraints can also benefit medical image segmentation and reconstruction tasks [20]. Furthermore, the prominent Medical Image Computing and Computer Assisted Intervention (MICCAI) society has established a special interest group in Shape in Medical Imaging (ShapeMI). This group is dedicated to exploring the applications of both traditional and contemporary (e.g., learning-based) shape analysis methods in medical imaging. Table 1 presents a partial list of professional organizations and events that are committed to this objective.
A non-inclusive list of organizations & events featuring shape and computer vision methods for medical applications.
| Sources | Description | Category |
|---|---|---|
| Zuse Institute Berlin (ZIB) | Shape-informed medical image segmentation and shape priors in medical imaging | Research group |
| ShapeMI | Shape processing/analysis/learning in medical imaging | MICCAI workshop |
| SIG | Shape modeling and analysis in medical imaging | MICCAI special interest group (SIG) |
| AutoImplant I, II | Skull shape reconstruction and completion | MICCAI challenge |
| WiSh | Women in shape analysis, shape modeling | Professional organization |
| STACOM | Statistical atlases and computational models of the heart | MICCAI workshop |
| SAMIA | Shape analysis in medical image analysis | Book |
| CIBC | Image and geometric analysis | Research group |
| GeoMedIA | Geometric deep learning in medical image analysis | MICCAI-endorsed workshop |
| IEEE TMI | Geometric deep learning in medical imaging | Journal special issue |
| PMLR | Geometric deep learning in medical image analysis | Proceedings |
| Elsevier | Riemannian geometric statistics in medical image analysis | Book |
| Springer | Geometric methods in bio-medical image processing | Proceedings |
| MCV | Workshop on medical computer vision | CVPR workshop |
| MCV 2010–2016 | Workshop on medical computer vision | MICCAI workshop |
| MeshMed | Workshop on mesh processing in medical image analysis | MICCAI workshop |
Nevertheless, state-of-the-art (SOTA) algorithms connot be directly applied to medical problems, since the vision methods were developed on general 3D shapes from ShapeNet and not on volumetric, gray-scale medical data. Therefore, the community needs a large, high-quality shape database for medical imaging that represents a variety of 3D medical shapes, i.e., voxel occupancy grid (VOR), mesh and point representations of human anatomies [21]. The inclusion of diverse anatomical shapes can aid in the development and evaluation of data-driven, shape-based methods for both vision and medical problems.
Computer vision methods, such as facial modeling [22] and internal anatomy inference [23] involve anatomical shapes, and medical problems can be solved using shape-based methods. Cranial implant design [24], [25], [26], [27], [28] is a typical example of a clinical problem that is commonly solved using well-established shape completion methods [29]. Such a shape completion concept can also be straightforwardly extended to other anatomical structures or even the whole body [30]. Therefore, there is a need for both normal and pathological anatomies to solve shape-based problems that are conventionally addressed using gray-scale medical images, e.g., extacting biomarkers [31].
In this paper, we present MedShapeNet, (1) a unique dataset for medical imaging shapes that serve complementary to existing shape benchmarks in computer vision, (2) a gap-bridger between the medical imaging and computer vision communities, and (3) a publicly available, continuous extending resource for benchmarking, education, extended reality (XR) applications [32], and the investigation of anatomical shape variations.
While existing datasets, such as ShapeNet are comprised of 3D computer-aided design (CAD) models of real-world objects (e.g., plane, car, chair, desk), MedShapeNet provides 3D shapes extracted from the imaging data of real patients including healthy as well as pathological subjects (Figure 1).

Example shapes in MedShapeNet, including various bones (e.g., skulls, ribs and vertebrae), organs (e.g., brain, lung, heart, liver), vessels (e.g., aortic vessel tree and pulmonary artery) and muscles.
Shape and voxel features
Shapes describe objects’ geometries, provide a foundation for computer vision, and serve as a computationally efficient way to represent images despite not capturing voxel features. In medicine, numerous diseases alter the morphological attributes of the affected anatomical structures (Figure 2). For instance, neoplastic formations, such as tumors, significant alter the morphologies of organs like the brain and the liver (Figure 3); neurological disorders, including Alzheimer’s disease (AD) [33], Parkinson’s disease (PD) [34] and substance use disorders, for instance, alcohol use disorder (AUD) and cocaine use disorder (CUD), can also cause morphological changes of brain substructures, such as the cerebral ventricles and the subcortical structures. These morphologic alterations allow disease detection and classification either manually, by medical professionals or automatically, through the application of specialized (e.g., shape analysis) machine learning algorithms.
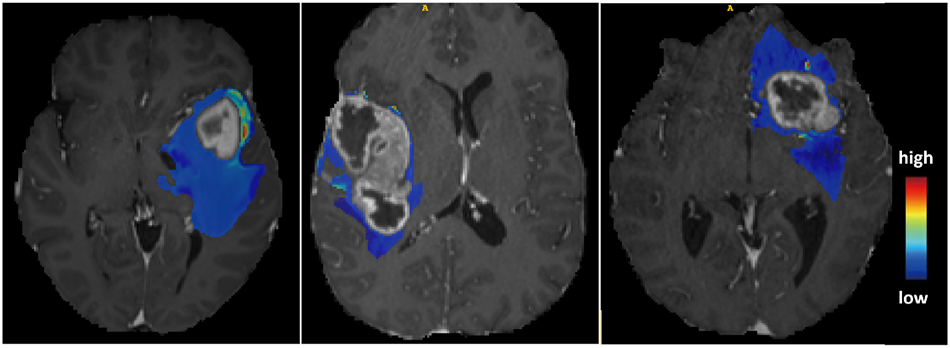
The predictive maps overlaid onto patients’ MRI scans. The predictive maps are color-coded to indicate high or low probability of tumor infiltration.

Example pathological shapes in MedShapeNet, including tumorous kidney (paired), brain (with real and synthetic tumors), liver and head & neck, as well as diseased coronary arteries. For illustration purpose, the opacity of some shapes is reduced to reveal the underlying tumors. We can study the effects of tumors on the morphological changes of an anatomy (e.g., brain) using such pathological data.
Hence, MedShapeNet highlights the significance of shape features, including jaggedness, volume, elongation, etc., over voxel features, such as intensities, for disease characterization, current medical image analysis tasks are still dominated by voxel-based methods. For instance, the so-called voxel-wise spatial predictive maps, as demonstrated by Akbari et al. [35], can pinpoint areas of early recurrence and infiltration of glioblastoma. These maps can be effectively used for targeted radiotherapy [36] (Figure 2), as regions with high probability are associated with a greater risk of tumor recurrence after resection. A naturally arising question is whether such predictive maps can be derived from the tumors’ geometries. MedShapeNet provides a platform to investigate the question and more:
What diseases can be comprehensively characterized by the shape features of the affected anatomical structures, and what diseases are solely reflected on voxel features?
How can one obtain discriminative shape features for disease detection using a machine learning model, either by handcrafting or learning them automatically using a deep network?
How to effectively combine shape and voxel features when shape features alone are insufficient for disease detection?
Do changes in voxel and shape features correlate statistically, and if so, how can this correlation be quantified?
Which of the current voxel-based mainstream approaches can be substituted with computationally more efficient shape-based methods for the analysis of medical data?
Transitioning from gray-scale imaging data to shape data and shape-based methods brings three primary benefits:
Shape data contain less identifying information than gray-scale imaging data, reducing the vulnerability to privacy attack when they are publicly shared [40];
Training on shape data encourages a deep network to concentrate on learning discriminative geometric features instead of patients’ identities irrelevant to the task. This can help improve the robustness and trustworthiness and prevent identity-driven bias of the learning system.
Sources of shapes
The shapes in MedShapeNet mostly originate from high-quality segmentation masks of anatomical structures, including different organs, bones, vessels, muscles, etc. (Figure 4). They are generated manually by domain experts, as those of the ground truth segmentations provided by medical image segmentation challenges [41], or semi-automatically, with the help of a segmentation network (e.g., TotalSegmentator [42], autoPET whole-body segmentation [43], AbdomenAtlas [44]). The majority of semi-automatic segmentations were also quality-checked by experts. Anatomical shapes with sophisticated geometric structures, such as the pulmonary trees (Figure 5), are also included in the MedShapeNet collection. In our terminology, we refer to binary voxel occupancy grids as segmentation masks, which we subsequently convert to meshes and point clouds using the Marching Cubes algorithm [45]. The majority of the source segmentation datasets are Creative Commons (CC) – licensed (Table 2), allowing us to adapt and redistribute the data. Furthermore, MedShapeNet includes both normal (Figure 1) and pathological shapes (Figure 3), delivered by the imaging data of healthy and diseased subjects, respectively. In addition, MedShapeNet provides 3D medical instrument models acquired using 3D handheld scanners [46] (Figure 4).

Illustration of 3D models of medical instruments used in oral and cranio-maxillofacial surgeries. The 3D models are obtained using structured light 3D scanners (Artec Leo from Artec3D and AutoScan Inspec from shining 3D). Instrument models can be retrieved by the search query instrument via the MedShapeNet web interface. Image taken from https://xrlab.ikim.nrw/.

Illustration of a pulmonary tree comprising the airway, artery and vein – thin structures that are difficult to segment and reconstruct.
The sources segmentation datasets (ordered alphabetically).
| Sources | Description | Dataset license |
|---|---|---|
| AbdomenAtlas [44] | 25 organs and seven types of tumor | – |
| AbdomenCT-1K [47] | Abdomen organs | CC BY 4.0 |
| AMOS [48] | Abdominal multi organs in CT and MRI | CC BY 4.0 |
| ASOCA [49], 50] | Normal and diseased coronary arteries | – |
| autoPET [43], 51]–53] | Whole-body segmentations | CC BY 4.0 |
| AVT [54] | Aortic vessel trees | CC BY 4.0 |
| BraTS [55]–57] | Brain tumor segmentation | – |
| Calgary-campinas [58] | Brain structure segmentations | – |
| Crossmoda [59], 60] | Brain tumor and cochlea segmentation | CC BY 4.0 |
| CT-ORG [61] | Multiple organ segmentation | CC0 1.0 |
| Digital body preservation [62] | 3D scans of anatomical specimens | – |
| EMIDEC [63], 64] | Normal and pathological (infarction) myocardium | CC BY NC SA 4.0 |
| Facial models [65] | Facial models for augmented reality | CC BY 4.0 |
| FLARE [47], 66]–68] | 13 abdomen organs | – |
| GLISRT [69]–71] | Brain structures | TCIA restricted |
| HCP [72] | Paired brain-skull extracted from MRIs | Data use terms |
| HECKTOR [73], 74] | Head and neck tumor segmentation | – |
| ISLES22 [75] | Ischemic stroke lesion segmentation | CC-BY-4.0 |
| KiTS21 [76] | Kidney and kidney tumor segmentation | MIT |
| LiTS [77] | Liver tumor segmentation | – |
| LNDb [78], 79] | Lung nodules | CC BY NC ND 4.0 |
| LUMIERE [80] | Longitudinal glioblastoma | CC BY NC |
| MUG500+ [81] | Healthy and craniotomy CT skulls | CC BY 4.0 |
| MRI GBM [82] | Brain and GBM extracted from MRIs | CC BY 4.0 |
| PROMISE [83] | Prostate MRI segmentation | – |
| PulmonaryTree [84] | Pulmonary airways, arteries and veins | CC BY 4.0 |
| SkullBreak [85] | Complete and artificially defected skulls | CC BY 4.0 |
| SkullFix [85] | Complete and artificially defected skulls | CC BY 4.0 |
| SUDMEX CONN [86] | Healthy and (cocaine use disorder) CUD brains | CC0 |
| TCGA-GBM [57] | Glioblastoma | – |
| 3D-COSI [46] | 3D medical instrument models | CC BY 4.0 |
| 3DTeethSeg [87], 88] | 3D teeth scan segmentation | CC BY NC ND 4.0 |
| ToothFairy [89], 90] | Inferior alveolar canal | CC BY SA |
| TotalSegmentator [42] | Various anatomical structures | CC BY 4.0 |
| VerSe [91] | Large scale vertebrae segmentation | CC BY 4.0 |
AbdomenAtlas
The dataset provides masks of 25 anatomical structures and seven types of tumors, derived from 5,195 CTs of 26 hospitals across eight countries [44]. These anatomical structures include the spleen, right kidney, left kidney, gall bladder, esophagus, liver, stomach, aorta, postcava, portal and splenic veins, pancreas, right and left adrenal glands, duodenum, hepatic vessel, right and left lungs, colon, intestine, rectum, bladder, prostate, left and right femur heads, and celiac trunk. Shape quality is ensured through manual annotations by medical professionals supported by a semi-automatic active learning procedure. The pathology-confirmed tumors include kidney, liver, pancreatic, hepatic vessel, lung, colon, and kidney cysts. The dataset provides a total of 51.8 K tumor masks. Moreover, a novel modeling-based tumor synthesis method is used to generates small, synthetic (<20 mm) tumor shapes [92], 93].
Pulmonary trees
The PulmonaryTree dataset [84] is a collection of pulmonary tree structures, amassed from 800 subjects across various medical centers in China [94]. It includes detailed 3D models of pulmonary airways, arteries, and veins, totaling 800 × 3=2,400 shapes. Each 3D model originates from CT scans with 512 × 512 voxels and 181–798 slices. The Z-spacing ranges from 0.5 to 1.5 mm. A collaborative annotation procedure ensures consistency provides a detailed and accurate representation of the pulmonary structures [95]. This procedure required approximately 3 h per case. The PulmonaryTree dataset introduces complex tree-like structures, a challenging aspect in medical image analysis (Figure 5). Specific technical challenges include maintaining the continuity of thin structures and addressing the uneven thickness of the main and branch structures.
TotalSegmentator
The dataset from Wasserthal et al. [42] includes over 1,000 CT scans and the masks of 104 anatomical structures covering the whole body. The masks are generated automatically by a nnUNet [96]. The data have been used to improve diagnosis by correlating organ volumes with disease occurrences [97].
Human connectome projects (HCP)
The 1,200 Subjects Data Release from HCP includes 1,113 structural 3T head MRI scans of healthy young adults. From each scan, the Cortical Surface Extraction script provided by BrainSuite [1] is used to extract the skull and brain masks.
MUG500+
This dataset contains the binary masks and meshes of 500 healthy human skulls and 29 craniectomy skulls with surgical defects [81]. Thresholding delivered the masks from head CT scans.
SkullBreak/SkullFix
The dataset includes the binary masks of healthy human skulls and the corresponding skulls with artificial defects. Similar to MUG500+ [81], thresholding head CTs from the CQ500 dataset [2] yields the masks.
Aortic vessel tree (AVT)
The dataset contains 56 computed tomography angiography (CTA) scans of healthy aortas and the masks of the aortic vessel trees [54], including the aorta, the aortic arch, the aortic branch, and the iliac arteries (Figure 1).
Vertebrae segmentation (VerSe)
The VerSe challenge provides the masks of vertebrae from around 210 subjects [91]. In total, 2,745 vertebra shapes are generated.
Automated segmentation of coronary arteries (ASOCA)
The ASOCA challenge provides the manual segmentations of 20 normal and 20 diseased coronary arteries [50].
3D teeth scan segmentation and labeling challenge (3DTeethSeg)
Automated teeth localization, segmentation, and labeling from intra-oral 3D scans significantly improve dental diagnostics, treatment planning, and population-based studies on oral health. Before initiating any orthodontic or restorative treatment, it is essential for a CAD system to accurately segment and label each instance of teeth. This eliminates the need of time-consuming manual adjustments by the dentist. The 3DTeethSeg provides the upper and lower jaw scans of 900 subjects, and the manual segmentations of the teeth, obtained from clinical evaluators with more than 10 years of expertise [87], 88].
Lung cancer patient management (LNDb) challenge
This dataset comprises lung nodule in low-dose CTs recorded for lung cancer screening [78], 79]. A total of 861 lung nodule masks correspond to 625 individual nodules segmented from 204 CTs. Five radiologists identified all pulmonary nodules with an in-plane dimension of 3 mm and higher.
Evaluation of myocardial infarction from delayed-enhancement cardiac MRI (EMIDEC)
This EMIDEC challenge provides 150 delayed enhancement MRI (DE-MRI) images in short axis orientation of the left ventricles. Experts contoured the myocardium and infarction areas in normal (50 cases) and pathological (100 cases) cases [63], 64]. The images were acquired roughly 10 min after the injection of a gadolinium-based contrast agent. The dataset is owned by the University Hospital of Dijon (France), but it is freely available.
ToothFairy
Placing dental implant can become complex when the implant hits the inferior alveolar nerve. The ToothFairy dataset contains cone-beam computed tomography (CBCT) images and was released for a segmentation challenge in 2023 [90]. It extends the previous datasets (i.e. [98]) and comprises 443 dental scans with a voxel size of 0.3 mm3 yielding volumes with shapes ranging from (148, 265, 312) to (169, 342, 370) across the Z, Y, and X axes, respectively. The dataset includes 2D sparse annotations for all 443 vol, while only a subset of 153 vol contains detailed 3D voxel-level annotations. A team of five experienced surgeons delivered the ground truth [99], 100]. Additionally, a test set of 50 CBCT with a voxel size of 0.4 mm3 is provided for evaluation.
HEad and neCK TumOR segmentation and outcome prediction (HECKTOR)
The training set of the HECKTOR challenge comprises 524 PET-CT volumes from seven hospitals with manual primary tumor and metastatic lymph nodes contours [73]. The data originates from FDG-PET and low-dose non-contrast-enhanced CT images of the head and neck region of subjects suffering from oropharyngeal cancer. The training set of the this challenge is provided to MedShapeNet.
autoPET
Similar to TotalSegmentor, whole-body segmentations are extracted from the PET-CT dataset provided by the autoPET challenge [51], using an semi-supervised segmentation network [43]. The dataset comes from cancer patients and includes manual masks of tumor lesions.
Calgary-campinas (CC)
This dataset provides high-quality anatomical data with 1 mm3 voxels from T1-weighted MRIs of 359 healthy subjects on scanners from three different vendors (GE, Philips, Siemens) at field strengths of 1.5 and 3 T [58]. The subjects vary in age and gender (176 M: 183 F, 53.5 ± 7.8 years, min: 18 years, max: 80 years). Probabilistic brain masks resulted from eight automated brain segmentation algorithms by simultaneous truth and performance level estimation (STAPLE) [101]. The quality of the masks was validated against 12 manual brain segmentations. Scientists investigate brain extraction models [102], domain shift and adaptation in brain MRI [103], as well as MRI reconstruction [104] using the CC dataset.
Abdominal multi-organ benchmark for segmentation (AMOS)
The AMOS data includes 500 CTs and 100 MRIs from a variety of scanners and locations [48]. It provides expert segmentations of 15 abdominal organs: spleen, right kidney, left kidney, gallbladder, esophagus, liver, stomach, aorta, inferior vena cava, pancreas, right adrenal gland, left adrenal gland, duodenum, bladder, and prostate/uterus. Patients with abdominal tumors or other abnormalities delivered the images.
AbdomenCT-1K and fast and low-resource abdominal organ segmentation (FLARE)
This dataset includes more than 1,000 CTs and manually generated masks of the liver, kidney, spleen, and pancreas [47]. A subset of the dataset was used in the [?] challenge, which provides expert segmentations of 13 abdomen organs the right and left kidney, stomach, gallbladder, esophagus, aorta, inferior vena cava, right adrenal gland, left adrenal gland, and duodenum [66] some of the CT scans are acquired from cancer patients.
Ischemic stroke lesion segmentation (ISLES)
The ISLES challenge [75] provides 250 brain MRIs with binary masks depicting stroke infarctions. The dataset encompasses diverse brain lesions in terms of volume, location, and stroke pattern. Masks are generated by manually refining automatic segmentations from a 3D UNet [105].
Synthetic anatomical shapes and shape augmentation
In addition to real anatomical shapes, we also provide synthetic shapes generated by generative adversarial net-works (GANs) [106]. For instance, we generate synthetic tumors for 27,390 real brains (Figure 3). Besides GANs, synthetic shapes can also be generated by registering two shapes and warping them to each other’s spaces [107]. This registration-based shape augmentation methods were used in the winning solutions of both the AutoImplant I and AutoImplant II challenges [26], 28].
Medical instruments
In addition to anatomical shapes, MedShapeNet also provides 3D models of medical instruments [46], such as drill bits, scalpels, and chisels (Figure 4). We process the structured-light 3D scans using proprietary software (Ultrascan 2.0.0.7, Artec Studio 17 Professional) to remove noise. These models could help develop surgical tool tracking methods in mixed reality for medical education and research [32]. Realistic and accurate virtual surgical planning is performed in AR or VR [108], which improves the surgical outcome [109].
Digital body preservation repository
These 3D models were captured from anatomical specimens using the handheld, high-resolution (accuracy 0.05 mm) structured-light surface scanner (Space Spider) and processed by the Studio 15 software (Artec 3D LUX, Luxembourg, Luxembourg) [62].
Pathological shapes
To increase the variability of the shape collections, MedShapeNet contains not only normal/healthy anatomical shapes, such as the kidneys from TotalSegmentor and the brains from HCP, but also pathological ones, which are derived from patients diagnosed with a specific pathological condition, such as tumor (liver, kidney, etc.) and CUD (SUDMEX CONN, Table 2). Figure 3 shows the tumorous kidneys, brains, livers and head & neck, as well as diseased coronary arteries from different sources. We also use generative adversarial networks (GANs) to generate synthetic brain tumors, as shown in Figure 3.
Annotation and example use cases
In MedShapeNet, pairedness is defined as having two composites i.e., the anatomical shapes and the metadata originating from the same subject, with one serving as input and the other as the ground truth. For instance, a 3D shape in MedShapeNet is paired with its anatomical category, such as ‘liver’, ‘heart’, ‘kidney’, and ‘lung’, which can be used for anatomical shape classification and retrieval. The metadata from DICOM or medical reports provides precise information about the source images, the patients (including attributes such as gender, age, body weight) as well as the diagnosis, and can deliver a variety of annotations. Synthetic shapes are distinguished from those obtained from real imaging data by the ‘synthetic’ label.
Benchmarks derived from MedShapeNet
From MedShapeNet and its paired data, we can derive three types of benchmark datasets (Table 3):
Discriminative benchmarks are comprised of 3D shapes and the corresponding anatomical categories and diagnosis. They can be used to train a classifier to discriminate 3D shapes (e.g., healthy, cancerous) based on shape-related features.
Reconstructive benchmarks are composed of anatomical shapes derived from whole-body segmentations. They can be used in shape reconstruction tasks. For example, by training on paired skull-face shapes (Figure 6(A)), we can reconstruct human faces from the skulls automatically. We can also estimate an individual’s body composition, such as fat percentage or muscle distribution from the body surface [110], 111], by regressing on paired skin-fat or skin-muscle data (Figure 6(C)), or create a missing organ from its surrounding anatomies [30].
Variational benchmarks are usually used for conditional reconstruction of 3D anatomical shapes. In addition to the geometric constraints imposed by the input shape, new reconstructions are expected to satisfy an additional attribute, such as age, gender or pathology. For example, it is possible to reconstruct multiple faces of different ages from the same skull, by introducing age as a constraint during supervised training. Similarly, a pathological condition, such as tumor, can be imposed on healthy anatomies, or the morphological changes of an anatomy during disease progression can be modeled [112]. Variational auto-encoder (VAE) [113] and GANs are commonly used for such conditional reconstruction tasks.
Instances of MedShapeNet benchmarks.
| Discriminative benchmarks | Reconstructive benchmarks | Variational benchmarks | |||
|---|---|---|---|---|---|
| Input (shape) | Ground truth (metadata) | Input (shape) | Ground truth (shape) | Input (shape + metadata) | Ground Truth (Shape) |
| Liver/kidney/brain | Tumor/healthy | Skull | Face | Face + AUD/CUD/AD/age | Face |
| Brain | AUD/CUD/AD/age | Ribs + spines | Torso organs | Brain + AUD/CUD/AD/age | Brain |
| Face | AUD/CUD/age/gender | Skin | Body fat/muscle/skeleton | – | – |
| 3D shapes | Anatomical categories | Full skeleton | Skin | – | – |
-
AD, Alzheimer’s disease; AUD, alcohol use disorder; CUD, cocaine use disorder.

Examples of paired anatomical shapes in MedShapeNet. (A) Paired skins, muscles, fat, different tissues, organs and bones. (B) Paired abdominal anatomies, including liver, spleen, pancreas, right kidney, left kidney, stomach, gallbladder, esophagus, aorta, inferior vena cava, right adrenal gland, left adrenal gland, and duodenum. (C) Paired internal anatomies and body surfaces. For anonymity, the faces are blurred.
Example use cases of MedShapeNet
To illustrate the unique value of MedShapeNet, we describe five real-world use cases and show how MedShapeNet is used to solve vision/medical problems:
Tumor classification of brain lesions is usually based on gray-scale MRIs [114], 115]. In this use case, we train a convolutional neural network (CNN)-based classifier to discriminate between tumorous and healthy brain shapes. The classifier has shown good convergence and generalizability. Similar results are observed for the classification of brain shapes from males and females, in line with existing studies [116].
Facial reconstruction is a common practice in archeology, anthropology and forensic science, where the objective is to recreate the facial appearances of historical figures, ancient humans or victims from their skeletal remains [117]. Orthognathic surgery also employs this technology to predict postoperative outcomes [118]. Nevertheless, in addition to the skull, the facial appearance is also significantly influenced by factors such as the quantity and distribution of facial fat and muscles [119], making facial reconstruction a highly ill-posed problem in terms of the skull-face relationship (Figure 7(A)).
Anatomy completion investigates the feasibility of automatically generating whole-body segmentations given only sparse manual annotations. The generated segmentations can subsequently be used as pseudo labels to train a whole-body segmentation network [30]. Figure 7(B) provides an example input and the corresponding reconstruction results.
Extended reality (XR) combines real and virtual worlds. MedShapeNet can also benefit a variety of XR (AR/MR/VR) applications that require 3D anatomical models [123], such as virtual anatomy education [124]. Figure 8(A) shows a whole-body model using the Microsoft HoloLens AR glasses. The user can dissemble individual anatomies, move them, zoom in and out, and rotate the structures (Figure 8(B) and (C)). Furthermore, if necessary, we can 3D print the models (Figure 8(D) and (E)). Users can also wear VR gloves (Figure 8(F)) to receive haptic feedback while interacting with the 3D anatomies in VR [125].

Benchmarks for various vision applications can be derived from MedShapeNet, such as (A) forensic facial reconstruction, (B) anatomical shape reconstruction, and (C) skull reconstruction.

A use case of MedShapeNet in AR- and VR-based anatomy education. (A) A whole-body model from MedShapeNet dissembled into individual anatomies. (B, C) anatomy manipulation in first- and third-person views. (D, E) A 3D-printed facial phantom and the corresponding skull and tumors. (F) Using haptic VR gloves to interact with the 3D anatomical models in the virtual environment.
MedShapeNet interface
Two interfaces are created for MedShapeNet, including a web-based interface that provides access to the original high-resolution shape data, and a Python API that enables users to interact with the shape data via Python.
Web-based interface
A user-centric, intuitive web-based interface[3] has been developed to provide convenient access to the shape data within MedShapeNet, which allows users to search, retrieve, and view individual shapes. Shapes can be retrieved using queries related to anatomical category such as heart’, brain, hip, liver, or pathologies like tumor. A dedicated GitHub page[4] has also been established to manage shape contribution and removal (in case of inaccurate shapes), feature requests and the open-sourcing of applications based on MedShapeNet.
MedShapeNetCore and python API
We have also developed a Python API that facilitates the integration of the dataset into Python-centric workflows for computer vision and machine learning. This API grants access to a standardized subset of the original MedShapeNet dataset, referred to as MedShapeNetCore, which has been specifically curated for the efficient and reliable benchmarking of various vision algorithms. MedShapeNetCore differs from the original dataset in aspects:
Quality. The 3D models in MedShapeNetCore are water-tight and the quality of each individual model has been meticulously verified through manual inspection.
Annotation. MedShapeNetCore is more densely annotated, expanding its applicability to tasks such as shape part segmentation [127] and anatomical symmetry plane estimation.
The 3D shapes are stored in the standard formats for geometric data structures, i.e., NIfTI (.nii) for voxel grids, stereolithography (.stl) for meshes and Polygon File Format (.ply) for point clouds, facilitating fast shape preview via existing softwares. The Python API facilitates the loading of these shape data into standard Numpy arrays, ensuring a seamless transformation into tensor representations compatible with various deep learning frameworks, including but not limited to PyTorch, MONAI, and TensorFlow. The light-weight nature of these data expedites the process of developing new medical vision algorithms or evaluating existing ones, while maintaining a low computational overhead. The ongoing efforts in the development of the Python API include integrating PyTorch3D [128] to leverage its sophisticated 3D operators, establishing predefined benchmarks tailored for various vision and medical applications, and incorporating pre-trained models and shape processing algorithms.
Discussion
High-quality, annotated datasets are valuable assets for data-driven research. We created MedShapeNet as an open, ongoing effort and requires continuous contributions from these communities. We believe that MedShapeNet holds the potential to make significant contributions to research in medical imaging and computer vision. It could impact the practice of medical data curation and sharing, as well as the development of data-driven methods for medical applications.
Compared to vision datasets, large medical datasets are more difficult to curate due to the sensitive, distributed, and scarce nature of medical images. Therefore, the medical imaging community has recently started catching up with the development of vision algorithms that can exploit large datasets, with more and more medical researchers becoming open to data-sharing. Thus, MedShapeNet provides a versatile dataset that both vision and medical researchers are accustomed to.
To avoid potentially harmful societal impact, computer vision research involving human-derived data should be conducted with care. We designed MedShapeNet specifically for research, and the researchers shall follow ethical guidelines throughout methodology development and experimental design. For example, publicly sharing neuroimaging data bears high privacy risks and needs regulation, since they contain patients’ facial profiles [129]. For instance, Schwarz et al. recently identified participants in a clinical trial comparing their faces reconstructed from MRI with photographs on social media [130]. Therefore, besides removing patients’ meta informationfrom DICOM tags, defacing is also commonly practiced [131]. However, we have shown that machine learning can reconstruct skulls even when they are damaged or parts of the bones are missing. Another double-edged use case of MedShapeNet is training machine learning to detect substance (drug or alcohol) addiction or other diseases e.g., fetal alcohol syndrome (FAS), based on facial characteristics [132]. Furthermore, since MedShapeNet preserves the correspondence between the shapes and patients’ meta information, such as age, race, gender, medical history, etc., which facilitates the learning of some controversial mapping relationships. Potentially, the ethnic identity or medical history is predicted from a person’s skull or facial profiles [133]. It is therefore the responsibility of the researchers to weigh the social benefits against the potential negative societal impacts while developing models using MedShapeNet.
For future developments, we will primarily focus on the following aspects:
Incorporating a greater number of datasets and metadata as well as pathological shapes, particularly those pertaining to rare diseases.
Advocating for MedShapeNet through presentations at conferences, symposia, and seminars, as well as organizing hackweeks, workshops, and challenges.
Establishing additional benchmarks and use cases.
Enhancing the web and Python interfaces.
Conclusions
In this white paper, we have introduced the initial efforts for MedShapeNet. We (1) formed a community for data contribution; (2) derived open-source benchmark datasets for several use cases; (3) constructed interfaces to search to download the shape data and its paired information; (4) brought up several interesting shape-related research topics; and (5) discussed the relevance of ethical guidelines and precautions for privacy of medical data.
Funding source: REACT-EU project KITE (Plattform für KI-Translation Essen, EFRE-0801977, https://kite.ikim.nrw/)
Funding source: FWF enFaced 2.0 (KLI 1044, https://enfaced2.ikim.nrw/), AutoImplant (https://autoimplant.ikim.nrw)
Funding source: “NUM 2.0” (FKZ: 01KX2121)
Funding source: Medical Faculty of RWTH Aachen University
Award Identifier / Grant number: Clinician Scientist Program
Funding source: National Natural Science Foundation of China
Award Identifier / Grant number: 81971709; M-0019; 82011530141
Funding source: Bundesministerium für Bildung und Forschung
Award Identifier / Grant number: BMBF, Ref. 161L0272
Funding source: Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen
Funding source: Der Regierende Bürgermeister von Berlin, Senatskanzlei Wissenschaft und Forschung
Funding source: Center for Virtual and Extended Reality in Medicine of the University Hospital Essen (ZvRM, https://zvrm.ume.de/)
Funding source: Cancer Imaging Archive (TCIA)
Funding source: National Cancer Institute
Award Identifier / Grant number: 1U01CA190214, 1U01CA187947
-
Research ethics: Not applicable.
-
Informed consent: Not applicable.
-
Author contributions: The authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Use of Large Language Models, AI and Machine Learning Tools: None declared.
-
Conflict of interest: All other authors state no conflict of interest.
-
Research funding: This work was supported by the REACT-EU project KITE (Plattform für KI-Translation Essen, EFRE-0801977, https://kite.ikim.nrw/), FWF enFaced 2.0 (KLI 1044, https://enfaced2.ikim.nrw/), AutoImplant (https://autoimplant.ikim.nrw/) and “NUM 2.0” (FKZ: 01KX2121). Behrus Puladi was funded by the Medical Faculty of RWTH Aachen University as part of the Clinician Scientist Program. In addition, we acknowledge the National Natural Science Foundation of China (81971709; M-0019; 82011530141). The work of J. Chen was supported by the Bundesministerium für Bildung und Forschung (BMBF, Ref. 161L0272). The work of ISAS was supported by the “Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen” and “Der Regierende Bürgermeister von Berlin, Senatskanzlei Wissenschaft und Forschung”. Furthermore, we acknowledge the Center for Virtual and Extended Reality in Medicine (ZvRM, https://zvrm.ume.de/) of the University Hospital Essen. The CT-ORG dataset was obtained from the Cancer Imaging Archive (TCIA). CT-ORG was supported in part by grants from the National Cancer Institute, 1U01CA190214 and 1U01CA187947. We thank all those who have contributed to the MedShapeNet collection (directly or indirectly).
-
Data availability: https://medshapenet.ikim.nrw/.
References
1. Esteva, A, Chou, K, Yeung, S, Naik, N, Madani, A, Mottaghi, A, et al.. Deep learning-enabled medical computer vision. npk Digital Med 2021;4:1–9. https://doi.org/10.1038/s41746-020-00376-2.Search in Google Scholar PubMed PubMed Central
2. Young, T, Hazarika, D, Poria, S, Cambria, E. Recent trends in deep learning based natural language processing. IEEE Comput Intell Mag 2018;13:55–75. https://doi.org/10.1109/mci.2018.2840738.Search in Google Scholar
3. Latif, S, Rana, R, Khalifa, S, Jurdak, R, Qadir, J, Schuller, BW. Deep representation learning in speech processing: challenges, recent advances, and future trends. arXiv preprint arXiv:2001.00378. 2020.Search in Google Scholar
4. Sun, C, Shrivastava, A, Singh, S, Gupta, A. Revisiting unreasonable effectiveness of data in deep learning era. In: Proceedings of the IEEE international conference on computer vision; 2017:843–52 pp.10.1109/ICCV.2017.97Search in Google Scholar
5. Egger, J, Gsaxner, C, Pepe, A, Pomykala, KL, Jonske, F, Kurz, M, et al.. Medical deep learning—a systematic meta-review. Comput Methods Progr Biomed 2022;221:106874. https://doi.org/10.1016/j.cmpb.2022.106874.Search in Google Scholar PubMed
6. Egger, J, Pepe, A, Gsaxner, C, Jin, Y, Li, J, Kern, R. Deep learning—a first meta-survey of selected reviews across scientific disciplines, their commonalities, challenges and research impact. PeerJ Comput Sci 2021;7:e773. https://doi.org/10.7717/peerj-cs.773.Search in Google Scholar PubMed PubMed Central
7. Deng, J, Dong, W, Socher, R, Li, L-J, Li, K, Fei-Fei, L. Imagenet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. Ieee; 2009:248–55 pp.10.1109/CVPR.2009.5206848Search in Google Scholar
8. Krizhevsky, A. Learning multiple layers of features from tiny images. 2009. University of Toronto, Report. https://www.cs.toronto.edu/~kriz/learning-features-2009-TR.pdf.Search in Google Scholar
9. Taylor, A, Marcus, M, Santorini, B. The penn treebank: an overview. In: Treebanks: building and using parsed corpora. Dordrecht: Springer Nature; 2003:5–22 pp.10.1007/978-94-010-0201-1_1Search in Google Scholar
10. Merity, S, Xiong, C, Bradbury, J, Socher, R. Pointer sentinel mixture models. arXiv preprint arXiv:1609.07843; 2016.Search in Google Scholar
11. Panayotov, V, Chen, G, Povey, D, Khudanpur, S. Librispeech: an ASR corpus based on public domain audio books. In: 2015 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE; 2015:5206–10 pp.10.1109/ICASSP.2015.7178964Search in Google Scholar
12. Wu, Z, Song, S, Khosla, A, Yu, F, Zhang, L, Tang, X, et al.. 3D shapenets: a deep representation for volumetric shapes. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2015:1912–20 pp.Search in Google Scholar
13. Chang, AX, Funkhouser, T, Guibas, L, Hanrahan, P, Huang, Q, Li, Z, et al.. Shapenet: an information-rich 3D model repository. arXiv preprint arXiv:1512.03012; 2015.Search in Google Scholar
14. Lin, M-X, Yang, J, Wang, H, Lai, Y-K, Jia, R, Zhao, B, et al.. Single image 3D shape retrieval via cross-modal instance and category contrastive learning. In: Proceedings of the IEEE/CVF international conference on computer vision; 2021:11405–15 pp.10.1109/ICCV48922.2021.01121Search in Google Scholar
15. Yan, X, Lin, L, Mitra, NJ, Lischinski, D, Cohen-Or, D, Huang, H. Shapeformer: transformer-based shape completion via sparse representation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2022:6239–49 pp.10.1109/CVPR52688.2022.00614Search in Google Scholar
16. Yi, L, Shao, L, Savva, M, Huang, H, Zhou, Y, Wang, Q, et al.. Large-scale 3d shape reconstruction and segmentation from shapenet core55. arXiv preprint arXiv:1710.06104; 2017.Search in Google Scholar
17. Sarasua, I, Pölsterl, S, Wachinger, C. Hippocampal representations for deep learning on alzheimer’s disease. Sci Rep 2022;12:8619. https://doi.org/10.1038/s41598-022-12533-6.Search in Google Scholar PubMed PubMed Central
18. Heimann, T, Meinzer, H-P. Statistical shape models for 3d medical image segmentation: a review. Med Image Anal 2009;13:543–63. https://doi.org/10.1016/j.media.2009.05.004.Search in Google Scholar PubMed
19. Petrelli, L, Pepe, A, Disanto, A, Gsaxner, C, Li, J, Jin, Y, et al.. Geometric modeling of aortic dissections through convolution surfaces. In: Medical imaging 2022: imaging informatics for healthcare, research, and applications, vol 12037. SPIE; 2022:198–206 pp.10.1117/12.2628187Search in Google Scholar
20. Yang, J, Wickramasinghe, U, Ni, B, Fua, P. Implicitatlas: learning deformable shape templates in medical imaging. In: CVPR. Danvers, MA, United States: IEEE; 2022:15861–71 pp.10.1109/CVPR52688.2022.01540Search in Google Scholar
21. Rezanejad, M, Khodadad, M, Mahyar, H, Lombaert, H, Gruninger, M, Walther, D, et al.. Medial spectral coordinates for 3D shape analysis. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2022:2686–96 pp.10.1109/CVPR52688.2022.00271Search in Google Scholar
22. Kania, K, Garbin, SJ, Tagliasacchi, A, Estellers, V, Yi, KM, Valentin, J, et al.. Blendfields: few-shot example-driven facial modeling. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2023:404–15 pp.10.1109/CVPR52729.2023.00047Search in Google Scholar
23. Keller, M, Zuffi, S, Black, MJ, Pujades, S. Osso: obtaining skeletal shape from outside. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2022:20492–501 pp.10.1109/CVPR52688.2022.01984Search in Google Scholar
24. Li, J, Pepe, A, Gsaxner, C, Campe, GV, Egger, J. A baseline approach for autoimplant: the miccai 2020 cranial implant design challenge. In: Workshop on clinical image-based procedures. Lima, Peru: Springer; 2020:75–84 pp.10.1007/978-3-030-60946-7_8Search in Google Scholar
25. Morais, A, Egger, J, Alves, V. Automated computer-aided design of cranial implants using a deep volumetric convolutional denoising autoencoder. In: World conference on information systems and technologies. Springer; 2019:151–60 pp.10.1007/978-3-030-16187-3_15Search in Google Scholar
26. Li, J, Pimentel, P, Szengel, A, Ehlke, M, Lamecker, H, Zachow, S, et al.. Autoimplant 2020-first miccai challenge on automatic cranial implant design. IEEE Trans Med Imag 2021;40:2329–42. https://doi.org/10.1109/tmi.2021.3077047.Search in Google Scholar PubMed
27. Li, J, von Campe, G, Pepe, A, Gsaxner, C, Wang, E, Chen, X, et al.. Automatic skull defect restoration and cranial implant generation for cranioplasty. Med Image Anal 2021;73:102171. https://doi.org/10.1016/j.media.2021.102171.Search in Google Scholar PubMed
28. Li, J, Ellis, DG, Kodym, O, Rauschenbach, L, Rieß, C, Sure, U, et al.. Towards clinical applicability and computational efficiency in automatic cranial implant design: an overview of the autoimplant 2021 cranial implant design challenge. Med Image Anal 2023:102865. https://doi.org/10.1016/j.media.2023.102865.Search in Google Scholar PubMed
29. Dai, A, Ruizhongtai Qi, C, Nießner, M. Shape completion using 3D-encoder-predictor CNNS and shape synthesis. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2017:5868–77 pp.10.1109/CVPR.2017.693Search in Google Scholar
30. Li, J, Pepe, A, Luijten, G, Schwarz-Gsaxner, C, Kleesiek, J, Egger, J. Anatomy completor: a multi-class completion framework for 3D anatomy reconstruction. arXiv preprint 2023. https://doi.org/10.1007/978-3-031-46914-5_1.Search in Google Scholar
31. Zhang, D, Huang, F, Khansari, M, Berendschot, TT, Xu, X, Dashtbozorg, B, et al.. Automatic corneal nerve fiber segmentation and geometric biomarker quantification. Eur Phys J Plus 2020;135:266. https://doi.org/10.1140/epjp/s13360-020-00127-y.Search in Google Scholar
32. Gsaxner, C, Li, J, Pepe, A, Schmalstieg, D, Egger, J. Inside-out instrument tracking for surgical navigation in augmented reality. In: Proceedings of the 27th ACM symposium on virtual reality software and technology; 2021:1–11 pp.10.1145/3489849.3489863Search in Google Scholar
33. Ohnishi, T, Matsuda, H, Tabira, T, Asada, T, Uno, M. Changes in brain morphology in alzheimer disease and normal aging: is alzheimer disease an exaggerated aging process? Am J Neuroradiol 2001;22:1680–5.Search in Google Scholar
34. Deng, J-H, Zhang, H-W, Liu, X-L, Deng, H-Z, Lin, F. Morphological changes in Parkinson’s disease based on magnetic resonance imaging: a mini-review of subcortical structures segmentation and shape analysis. World J Psychiatr 2022;12:1356. https://doi.org/10.5498/wjp.v12.i12.1356.Search in Google Scholar PubMed PubMed Central
35. Akbari, H, Macyszyn, L, Da, X, Bilello, M, Wolf, RL, Martinez-Lage, M, et al.. Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery 2016;78:572. https://doi.org/10.1227/neu.0000000000001202.Search in Google Scholar
36. Seker-Polat, F, Pinarbasi Degirmenci, N, Solaroglu, I, Bagci-Onder, T. Tumor cell infiltration into the brain in glioblastoma: from mechanisms to clinical perspectives. Cancers 2022;14:443. https://doi.org/10.3390/cancers14020443.Search in Google Scholar PubMed PubMed Central
37. Li, J, Gsaxner, C, Pepe, A, Schmalstieg, D, Kleesiek, J, Egger, J. Sparse convolutional neural network for high-resolution skull shape completion and shape super-resolution. Sci Rep 2023;13. https://doi.org/10.1038/s41598-023-47437-6.Search in Google Scholar PubMed PubMed Central
38. Jin, L, Gu, S, Wei, D, Adhinarta, JK, Kuang, K, Zhang, YJ, et al.. Ribseg v2: a large-scale benchmark for rib labeling and anatomical centerline extraction. IEEE Trans Med Imag 2023. https://doi.org/10.1109/tmi.2023.3313627.Search in Google Scholar
39. Wickramasinghe, U, Jensen, P, Shah, M, Yang, J, Fua, P. Weakly supervised volumetric image segmentation with deformed templates. In: MICCAI. Singapore: Springer; 2022:422–32 pp.10.1007/978-3-031-16443-9_41Search in Google Scholar
40. De Kok, JW, Á, M, De la Hoz, A, de Jong, Y, Brokke, V, Elbers, PW, et al.. A guide to sharing open healthcare data under the general data protection regulation. Sci Data 2023;10:404. https://doi.org/10.1038/s41597-023-02256-2.Search in Google Scholar PubMed PubMed Central
41. Eisenmann, M, Reinke, A, Weru, V, Tizabi, MD, Isensee, F, Adler, T, et al.. Why is the winner the best? In: Proceedings of the IEEE/ CVF computer vision and pattern recognition conference (CVPR). IEEE; 2023.Search in Google Scholar
42. Wasserthal, J, Breit, H-C, Meyer, MT, Pradella, M, Hinck, D, Sauter, AW, et al.. Totalsegmentator: robust segmentation of 104 anatomical structures in CT images. Radiol Artif Intell 2023;5. https://doi.org/10.1148/ryai.230024.Search in Google Scholar PubMed PubMed Central
43. Jaus, A, Seibold, C, Hermann, K, Walter, A, Giske, K, Haubold, J, et al.. Towards unifying anatomy segmentation: automated generation of a full-body CT dataset via knowledge aggregation and anatomical guidelines. arXiv preprint arXiv:2307.13375; 2023.10.1109/ICIP51287.2024.10647307Search in Google Scholar
44. Qu, C, Zhang, T, Qiao, H, Liu, J, Tang, Y, Yuille, A, et al.. Abdomenatlas-8k: annotating 8,000 abdominal CT volumes for multi-organ segmentation in three weeks. In: Conference on neural information processing systems; 2023.Search in Google Scholar
45. Lorensen, WE, Cline, HE. Marching cubes: a high resolution 3d surface construction algorithm. ACM SIGGRAPH Comput Graph 1987;21:163–9. https://doi.org/10.1145/37402.37422.Search in Google Scholar
46. Luijten, G, Gsaxner, C, Li, J, Pepe, A, Ambigapathy, N, Kim, M, et al.. 3D surgical instrument collection for computer vision and extended reality. Sci Data 2023;10. https://doi.org/10.1038/s41597-023-02684-0.Search in Google Scholar PubMed PubMed Central
47. Ma, J, Zhang, Y, Gu, S, Zhu, C, Ge, C, Zhang, Y, et al.. Abdomenct-1k: is abdominal organ segmentation a solved problem? IEEE Trans Pattern Anal Mach Intell 2022;44:6695–714. https://doi.org/10.1109/tpami.2021.3100536.Search in Google Scholar PubMed
48. Ji, Y., Bai, H., Ge, C., Yang, J., Zhu, Y., Zhang, R., Li, Z., Zhanng, L., Ma, W., Wan, X., et al.. Amos: A large-scale abdominal multi-organ benchmark for versatile medical image segmentation. In: Advances in neural information processing systems. NY, US: ACM Red Hook; 2022, 35:36722–32 pp.Search in Google Scholar
49. Gharleghi, R, Adikari, D, Ellenberger, K, Ooi, S-Y, Ellis, C, Chen, C-M, et al.. Automated segmentation of normal and diseased coronary arteries – the asoca challenge. Comput Med Imag Graph 2022;97:102049. https://doi.org/10.1016/j.compmedimag.2022.102049.Search in Google Scholar PubMed
50. Gharleghi, R, Adikari, D, Ellenberger, K, Webster, M, Ellis, C, Sowmya, A, et al.. Annotated computed tomography coronary angiogram images and associated data of normal and diseased arteries. Sci Data 2023;10:128. https://doi.org/10.1038/s41597-023-02016-2.Search in Google Scholar PubMed PubMed Central
51. Gatidis, S, Hepp, T, Früh, M, La Fougère, C, Nikolaou, K, Pfannenberg, C, et al.. A whole-body FDG-PET/CT dataset with manually annotated tumor lesions. Sci Data 2022;9:601. https://doi.org/10.1038/s41597-022-01718-3.Search in Google Scholar PubMed PubMed Central
52. Gatidis, S, Früh, M, Fabritius, M, Gu, S, Nikolaou, K, La Fougère, C, et al.. Results from the autoPET challenge on fully automated lesion segmentation in oncologic PET/CT imaging. Nat Mach Intell 2024;1–20. https://doi.org/10.21203/rs.3.rs-2572595/v1.Search in Google Scholar
53. Gatidis, S, Küstner, T, Früh, M, La Fougère, C, Nikolaou, K, Pfannenberg, C, et al.. A whole-body FDG-PET/CT dataset with manually annotated tumor lesions. Cancer Imag Arch 2022. https://doi.org/10.7937/gkr0-xv29.Search in Google Scholar
54. Radl, L, Jin, Y, Pepe, A, Li, J, Gsaxner, C, Zhao, F-H., et al.. Avt: multicenter aortic vessel tree CTA dataset collection with ground truth segmentation masks. Data in Brief 2022;40:107801. https://doi.org/10.1016/j.dib.2022.107801.Search in Google Scholar PubMed PubMed Central
55. Baid, U, Ghodasara, S, Mohan, S, Bilello, M, Calabrese, E, Colak, E, et al.. The RSNA-ASNR-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv preprint arXiv:2107.02314; 2021.Search in Google Scholar
56. Menze, BH, Jakab, A, Bauer, S, Kalpathy-Cramer, J, Farahani, K, Kirby, J, et al.. The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans Med Imag 2014;34:1993–2024. https://doi.org/10.1109/tmi.2014.2377694.Search in Google Scholar
57. Bakas, S, Akbari, H, Sotiras, A, Bilello, M, Rozycki, M, Kirby, JS, et al.. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4:1–13. https://doi.org/10.1038/sdata.2017.117.Search in Google Scholar PubMed PubMed Central
58. Souza, R, Lucena, O, Garrafa, J, Gobbi, D, Saluzzi, M, Appenzeller, S, et al.. An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. Neuroimage 2018;170:482–94. https://doi.org/10.1016/j.neuroimage.2017.08.021.Search in Google Scholar PubMed
59. Shapey, J, Kujawa, A, Dorent, R, Wang, G, Dimitriadis, A, Grishchuk, D, et al.. Segmentation of vestibular schwannoma from MRI, an open annotated dataset and baseline algorithm. Sci Data 2021;8:286. https://doi.org/10.1038/s41597-021-01064-w.Search in Google Scholar PubMed PubMed Central
60. Dorent, R, Kujawa, A, Ivory, M, Bakas, S, Rieke, N, Joutard, S, et al.. Crossmoda 2021 challenge: benchmark of cross-modality domain adaptation techniques for vestibular schwannoma and cochlea segmentation. Med Image Anal 2023;83:102628. https://doi.org/10.1016/j.media.2022.102628.Search in Google Scholar PubMed PubMed Central
61. Rister, B, Yi, D, Shivakumar, K, Nobashi, T, Rubin, DL. CT-ORG, a new dataset for multiple organ segmentation in computed tomography. Sci Data 2020;7:381. https://doi.org/10.1038/s41597-020-00715-8.Search in Google Scholar PubMed PubMed Central
62. Vandenbossche, V, Van de Velde, J, Avet, S, Willaert, W, Soltvedt, S, Smit, N, et al.. Digital body preservation: technique and applications. Anat Sci Educ 2022;15:731–44. https://doi.org/10.1002/ase.2199.Search in Google Scholar PubMed
63. Lalande, A, Chen, Z, Decourselle, T, Qayyum, A, Pommier, T, Lorgis, L, et al.. Emidec: a database useable for the automatic evaluation of myocardial infarction from delayed-enhancement cardiac MRI. Data 2020;5:89. https://doi.org/10.3390/data5040089.Search in Google Scholar
64. Lalande, A, Chen, Z, Pommier, T, Decourselle, T, Qayyum, A, Salomon, M, et al.. Deep learning methods for automatic evaluation of delayed enhancement-mri. The results of the emidec challenge. Med Image Anal 2022;79:102428. https://doi.org/10.1016/j.media.2022.102428.Search in Google Scholar PubMed
65. Gsaxner, C, Wallner, J, Chen, X, Zemann, W, Egger, J. Facial model collection for medical augmented reality in oncologic cranio-maxillofacial surgery. Sci Data 2019;6:1–7. https://doi.org/10.1038/s41597-019-0327-8.Search in Google Scholar PubMed PubMed Central
66. Ma, J, Zhang, Y, Gu, S, An, X, Wang, Z, Ge, C, et al.. Fast and low-GPU-memory abdomen CT organ segmentation: the flare challenge. Med Image Anal 2022;82:102616. https://doi.org/10.1016/j.media.2022.102616.Search in Google Scholar PubMed
67. Simpson, AL, Antonelli, M, Bakas, S, Bilello, M, Farahani, K, Van Ginneken, B, et al.. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063; 2019.Search in Google Scholar
68. Ma, J., Zhang, Y., Gu, S., Ge, C., Ma, S., Young, A., et al.. Unleashing the strengths of unlabeled data in pan-cancer abdominal organ quantification: the flare22 challenge. arXiv preprint arXiv:2308.05862; 2023.10.1016/S2589-7500(24)00154-7Search in Google Scholar PubMed
69. Shusharina, N, Bortfeld, T. Glioma image segmentation for radiotherapy: RT targets, barriers to cancer spread, and organs at risk (GLIS-RT). Cancer Imag Arch 2021. https://doi.org/10.7937/TCIA.T905-ZQ20.Search in Google Scholar
70. Shusharina, N, Bortfeld, T, Cardenas, C, De, B, Diao, K, Hernandez, S, et al.. Cross-modality brain structures image segmentation for the radiotherapy target definition and plan optimization. In: Segmentation, classification, and registration of multi-modality medical imaging data: MICCAI 2020 challenges, ABCs 2020, L2R 2020, TN-SCUI 2020, held in conjunction with MICCAI 2020, Lima, Peru, October 4–8, 2020, proceedings 23. Springer; 2021:3–15 pp.10.1007/978-3-030-71827-5_1Search in Google Scholar
71. Shusharina, N, Söderberg, J, Edmunds, D, Löfman, F, Shih, H, Bortfeld, T. Automated delineation of the clinical target volume using anatomically constrained 3D expansion of the gross tumor volume. Radiother Oncol 2020;146:37–43. https://doi.org/10.1016/j.radonc.2020.01.028.Search in Google Scholar PubMed PubMed Central
72. Elam, JS, Glasser, MF, Harms, MP, Sotiropoulos, SN, Andersson, JL, Burgess, GC, et al.. The human connectome project: a retrospective. Neuroimage 2021;244:118543. https://doi.org/10.1016/j.neuroimage.2021.118543.Search in Google Scholar PubMed PubMed Central
73. Andrearczyk, V, Oreiller, V, Abobakr, M, Akhavanallaf, A, Balermpas, P, Boughdad, S, et al.. Overview of the HECKTOR challenge at MICCAI 2022: automatic head and neck tumor segmentation and outcome prediction in PET/CT. In: Head and neck tumor segmentation and outcome prediction. Singapore: Springer; 2022:1–30 pp.10.1007/978-3-031-27420-6_1Search in Google Scholar PubMed PubMed Central
74. Oreiller, V, Andrearczyk, V, Jreige, M, Boughdad, S, Elhalawani, H, Castelli, J, et al.. Head and neck tumor segmentation in PET/CT: the hecktor challenge. Med Image Anal 2022;77:102336. https://doi.org/10.1016/j.media.2021.102336.Search in Google Scholar PubMed
75. Hernandez Petzsche, MR, de la Rosa, E, Hanning, U, Wiest, R, Valenzuela, W, Reyes, M, et al.. ISLES 2022: a multi-center magnetic resonance imaging stroke lesion segmentation dataset. Sci Data 2022;9:762. https://doi.org/10.1038/s41597-022-01875-5.Search in Google Scholar PubMed PubMed Central
76. Heller, N, Isensee, F, Maier-Hein, KH, Hou, X, Xie, C, Li, F, et al.. The state of the art in kidney and kidney tumor segmentation in contrast-enhanced ct imaging: results of the KITS19 challenge. Med Image Anal 2020:101821. https://doi.org/10.1016/j.media.2020.101821.Search in Google Scholar PubMed PubMed Central
77. Bilic, P, Christ, P, Li, HB, Vorontsov, E, Ben-Cohen, A, Kaissis, G, et al.. The liver tumor segmentation benchmark (lits). Med Image Anal 2023;84:102680. https://doi.org/10.1016/j.media.2022.102680.Search in Google Scholar PubMed PubMed Central
78. Pedrosa, J, Aresta, G, Ferreira, C, Rodrigues, M, Leitão, P, Carvalho, AS, et al.. LNDb: a lung nodule database on computed tomography. arXiv preprint arXiv:1911.08434; 2019.Search in Google Scholar
79. Pedrosa, J, Aresta, G, Ferreira, C, Atwal, G, Phoulady, HA, Chen, X, et al.. LNDb challenge on automatic lung cancer patient management. Med Image Anal 2021;70:102027. https://doi.org/10.1016/j.media.2021.102027.Search in Google Scholar PubMed
80. Suter, Y, Knecht, U, Valenzuela, W, Notter, M, Hewer, E, Schucht, P, et al.. The lumiere dataset: longitudinal glioblastoma MRI with expert rano evaluation. Sci Data 2022;9:768. https://doi.org/10.1038/s41597-022-01881-7.Search in Google Scholar PubMed PubMed Central
81. Li, J, Krall, M, Trummer, F, Memon, AR, Pepe, A, Gsaxner, C, et al.. Mug500+: database of 500 high-resolution healthy human skulls and 29 craniotomy skulls and implants. Data Brief 2021;39:107524. https://doi.org/10.1016/j.dib.2021.107524.Search in Google Scholar PubMed PubMed Central
82. L Lindner, D Wild, M Weber, M Kolodziej, G von Campe, and J Egger, Skull-stripped MRI GBM datasets (and segmentations), 6 2019. https://figshare.com/articles/dataset/Skull-stripped_MRI_GBM_Datasets_and_Segmentations_/7435385.Search in Google Scholar
83. Litjens, G, Toth, R, Van De Ven, W, Hoeks, C, Kerkstra, S, Van Ginneken, B, et al.. Evaluation of prostate segmentation algorithms for MRI: the promise12 challenge. Med Image Anal 2014;18:359–73. https://doi.org/10.1016/j.media.2013.12.002.Search in Google Scholar PubMed PubMed Central
84. Weng, Z, Yang, J, Liu, D, Cai, W. Topology repairing of disconnected pulmonary airways and vessels: baselines and a dataset. In: MICCAI. Vancouver: Springer; 2023.10.1007/978-3-031-43990-2_36Search in Google Scholar
85. Kodym, O, Li, J, Pepe, A, Gsaxner, C, Chilamkurthy, S, Egger, J, et al.. Skullbreak/skullfix–dataset for automatic cranial implant design and a benchmark for volumetric shape learning tasks. Data Brief 2021;35:106902. https://doi.org/10.1016/j.dib.2021.106902.Search in Google Scholar PubMed PubMed Central
86. Angeles-Valdez, D, Rasgado-Toledo, J, Issa-Garcia, V, Balducci, T, Villicaña, V, Valencia, A, et al.. The mexican magnetic resonance imaging dataset of patients with cocaine use disorder: SUDMEX CONN. Sci Data 2022;9:133. https://doi.org/10.1038/s41597-022-01251-3.Search in Google Scholar PubMed PubMed Central
87. Ben-Hamadou, A, Smaoui, O, Rekik, A, Pujades, S, Boyer, E, Lim, H, et al.. 3DTeethSeg’22: 3D teeth scan segmentation and labeling challenge. arXiv preprint arXiv:2305.18277; 2023.Search in Google Scholar
88. Ben-Hamadou, A, Smaoui, O, Chaabouni-Chouayakh, H, Rekik, A, Pujades, S, Boyer, E, et al.. Teeth3Ds: a benchmark for teeth segmentation and labeling from intra-oral 3D scans. arXiv preprint arXiv:2210.06094; 2022.Search in Google Scholar
89. Cipriano, M, Allegretti, S, Bolelli, F, Pollastri, F, Grana, C. Improving segmentation of the inferior alveolar nerve through deep label propagation. In: IEEE/CVF conference on computer vision and pattern recognition (CVPR). IEEE; 2022:21 137–21 146 pp.10.1109/CVPR52688.2022.02046Search in Google Scholar
90. Bolelli, F, Lumetti, L, Di Bartolomeo, M, Vinayahalingam, S, Anesi, A, van Ginneken, B, et al.. Tooth fairy: a cone-beam computed tomography segmentation challenge. In: Structured challenge; 2023.Search in Google Scholar
91. Sekuboyina, A, Rempfler, M, Valentinitsch, A, Menze, BH, Kirschke, JS. Labeling vertebrae with two-dimensional reformations of multidetector CT images: an adversarial approach for incorporating prior knowledge of spine anatomy. Radiol Artif Intell 2020;2:e190074. https://doi.org/10.1148/ryai.2020190074.Search in Google Scholar PubMed PubMed Central
92. Hu, Q, Chen, Y, Xiao, J, Sun, S, Chen, J, Yuille, AL, et al.. Label-free liver tumor segmentation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2023:7422–32 pp.10.1109/CVPR52729.2023.00717Search in Google Scholar
93. Li, B, Chou, Y-C, Sun, S, Qiao, H, Yuille, A, Zhou, Z. Early detection and localization of pancreatic cancer by label-free tumor synthesis. In: MICCAI workshop on big task small data, 1001-AI; 2023.Search in Google Scholar
94. Kuang, K, Zhang, L, Li, J, Li, H, Chen, J, Du, B, et al.. What makes for automatic reconstruction of pulmonary segments. In: MICCAI. Singapore: Springer; 2022:495–505 pp.10.1007/978-3-031-16431-6_47Search in Google Scholar
95. Xie, K, Yang, J, Wei, D, Weng, Z, Fua, P. Efficient anatomical labeling of pulmonary tree structures via implicit point-graph networks. arXiv preprint arXiv:2309.17329; 2023.Search in Google Scholar
96. Isensee, F, Jaeger, PF, Kohl, SA, Petersen, J, Maier-Hein, KH. NNU-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203–11. https://doi.org/10.1038/s41592-020-01008-z.Search in Google Scholar PubMed
97. van Meegdenburg, T, Kleesiek, J, Egger, J, Perrey, S. Improvement in disease diagnosis in computed tomography images by correlating organ volumes with disease occurrences in humans. BioMedInformatics 2023;3:526–42. https://doi.org/10.3390/biomedinformatics3030036.Search in Google Scholar
98. Di Bartolomeo, M, Pellacani, A, Bolelli, F, Cipriano, M, Lumetti, L, Negrello, S, et al.. Inferior alveolar canal automatic detection with deep learning CNNs on CBCTs: development of a novel model and Release of open-source dataset and algorithm. Appl Sci 2023;13. https://doi.org/10.3390/app13053271.Search in Google Scholar
99. Lumetti, L, Pipoli, V, Bolelli, F, Grana, C. Annotating the inferior alveolar canal: the ultimate tool. In: Image analysis and processing – ICIAP 2023. Udine: Springer; 2023:1–12 pp.10.1007/978-3-031-43148-7_44Search in Google Scholar
100. Mercadante, C, Cipriano, M, Bolelli, F, Pollastri, F, Di Bartolomeo, M, Anesi, A, et al.. A cone beam computed tomography annotation tool for automatic detection of the inferior alveolar nerve canal. In: Proceedings of the 16th international joint conference on computer vision, imaging and computer graphics theory and applications – volume 4: VISAPP. SciTePress; 2021, 4:724–31 pp.10.5220/0010392307240731Search in Google Scholar
101. Warfield, SK, Zou, KH, Wells, WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imag 2004;23:903–21. https://doi.org/10.1109/tmi.2004.828354.Search in Google Scholar PubMed PubMed Central
102. Lucena, O, Souza, R, Rittner, L, Frayne, R, Lotufo, R. Convolutional neural networks for skull-stripping in brain mr imaging using silver standard masks. Artif Intell Med 2019;98:48–58. https://doi.org/10.1016/j.artmed.2019.06.008.Search in Google Scholar PubMed
103. Saat, P, Nogovitsyn, N, Hassan, MY, Ganaie, MA, Souza, R, Hemmati, H. A domain adaptation benchmark for t1-weighted brain magnetic resonance image segmentation. Front Neuroinf 2022:96. https://doi.org/10.3389/fninf.2022.919779.Search in Google Scholar PubMed PubMed Central
104. Yiasemis, G, Sonke, J-J, Sa´nchez, C, Teuwen, J. Recurrent variational network: a deep learning inverse problem solver applied to the task of accelerated MRI reconstruction. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2022:732–41 pp.10.1109/CVPR52688.2022.00081Search in Google Scholar
105. Öçiçek, Abdulkadir, A, Lienkamp, SS, Brox, T, Ronneberger, O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Medical image computing and computer-assisted intervention–MICCAI 2016: 19th international conference, Athens, Greece, October 17–21, 2016, proceedings, part II 19. Springer; 2016:424–32 pp.10.1007/978-3-319-46723-8_49Search in Google Scholar
106. Ferreira, A, Li, J, Pomykala, KL, Kleesiek, J, Alves, V, Egger, J. Gan-based generation of realistic 3D data: a systematic review and taxonomy. arXiv preprint arXiv:2207.01390; 2022.Search in Google Scholar
107. Ellis, DG, Aizenberg, MR. Deep learning using augmentation via registration: 1st place solution to the autoimplant 2020 challenge. In: Towards the automatization of cranial implant design in cranioplasty: first challenge, autoimplant 2020, held in conjunction with MICCAI 2020, Lima, Peru, October 8, 2020, proceedings 1. Springer; 2020:47–55 pp.10.1007/978-3-030-64327-0_6Search in Google Scholar
108. Velarde, K, Cafino, R, Isla, AJr, Ty, KM, Palmer, X-L, Potter, L, et al.. Virtual surgical planning in craniomaxillofacial surgery: a structured review. Comput Assist Surg 2023;28:2271160. https://doi.org/10.1080/24699322.2023.2271160.Search in Google Scholar PubMed
109. Laskay, NM, George, JA, Knowlin, L, Chang, TP, Johnston, JM, Godzik, J. Optimizing surgical performance using preoperative virtual reality planning: a systematic review. World J Surg 2023:1–11. https://doi.org/10.1007/s00268-023-07064-8.Search in Google Scholar PubMed
110. Mueller, TT, Zhou, S, Starck, S, Jungmann, F, Ziller, A, Aksoy, O, et al.. Body fat estimation from surface meshes using graph neural networks. In: International workshop on shape in medical imaging. Springer; 2023:105–17 pp.10.1007/978-3-031-46914-5_9Search in Google Scholar
111. Piecuch, L, Gonzales Duque, V, Sarcher, A, Hollville, E, Nordez, A, Rabita, G, et al.. Muscle volume quantification: guiding transformers with anatomical priors. In: International workshop on shape in medical imaging. Springer; 2023:173–87 pp.10.1007/978-3-031-46914-5_14Search in Google Scholar
112. Sauty, B, Durrleman, S. Progression models for imaging data with longitudinal variational auto encoders. In: International conference on medical image computing and computer-assisted intervention. Springer; 2022:3–13 pp.10.1007/978-3-031-16431-6_1Search in Google Scholar
113. Kingma, DP, Welling, M. Auto-encoding variational bayes. arXiv preprint arXiv:1312.6114; 2013.Search in Google Scholar
114. Amin, J, Sharif, M, Raza, M, Saba, T, Anjum, MA. Brain tumor detection using statistical and machine learning method. Comput Methods Progr Biomed 2019;177:69–79. https://doi.org/10.1016/j.cmpb.2019.05.015.Search in Google Scholar PubMed
115. Amin, J, Sharif, M, Haldorai, A, Yasmin, M, Nayak, RS. Brain tumor detection and classification using machine learning: a comprehensive survey. Complex Intell Syst 2021:1–23. https://doi.org/10.1007/s40747-021-00563-y.Search in Google Scholar
116. Xin, J, Zhang, Y, Tang, Y, Yang, Y. Brain differences between men and women: evidence from deep learning. Front Neurosci 2019;13:185. https://doi.org/10.3389/fnins.2019.00185.Search in Google Scholar PubMed PubMed Central
117. Missal, S. Forensic facial reconstruction of skeletonized and highly decomposed human remains. In: Forensic genetic approaches for identification of human skeletal remains. London: Elsevier; 2023:549–69 pp.10.1016/B978-0-12-815766-4.00026-1Search in Google Scholar
118. Lampen, N, Kim, D, Xu, X, Fang, X, Lee, J, Kuang, T, et al.. Spatiotemporal incremental mechanics modeling of facial tissue change. In: International conference on medical image computing and computer-assisted intervention. Springer; 2023:566–75 pp.10.1007/978-3-031-43996-4_54Search in Google Scholar
119. Damas, S, Cordón, O, Ibáñez, O. Relationships between the skull and the face for forensic craniofacial superimposition. In: Handbook on craniofacial superimposition: The MEPROCS project. Cham: Springer; 2020:11–50 pp.10.1007/978-3-319-11137-7_3Search in Google Scholar
120. Li, J, Fragemann, J, Ahmadi, S-A, Kleesiek, J, Egger, J. Training β-vae by aggregating a learned Gaussian posterior with a decoupled decoder. In: MICCAI workshop on medical applications with disentanglements. Springer; 2022:70–92 pp.10.1007/978-3-031-25046-0_7Search in Google Scholar
121. Friedrich, P, Wolleb, J, Bieder, F, Thieringer, FM, Cattin, PC. Point cloud diffusion models for automatic implant generation. In: International conference on medical image computing and computer-assisted intervention. Springer; 2023:112–22 pp.10.1007/978-3-031-43996-4_11Search in Google Scholar
122. Wodzinski, M, Daniol, M, Hemmerling, D, Socha, M. High-resolution cranial defect reconstruction by iterative, low-resolution, point cloud completion transformers. In: International conference on medical image computing and computer-assisted intervention. Springer; 2023:333–43 pp.10.1007/978-3-031-43996-4_32Search in Google Scholar
123. Gsaxner, C, Li, J, Pepe, A, Jin, Y, Kleesiek, J, Schmalstieg, D, et al.. The hololens in medicine: a systematic review and taxonomy. Med Image Anal 2023:102757. https://doi.org/10.1016/j.media.2023.102757.Search in Google Scholar PubMed
124. Bölek, KA, De Jong, G, Henssen, D. The effectiveness of the use of augmented reality in anatomy education: a systematic review and meta-analysis. Sci Rep 2021;11:15292. https://doi.org/10.1038/s41598-021-94721-4.Search in Google Scholar PubMed PubMed Central
125. Krieger, K, Egger, J, Kleesiek, J, Gunzer, M, Chen, J. Multimodal extended reality applications offer benefits for volumetric biomedical image analysis in research and medicine. arXiv preprint arXiv:2311.03986; 2023.10.1007/s10278-024-01094-xSearch in Google Scholar PubMed PubMed Central
126. Yang, J, Shi, R, Wei, D, Liu, Z, Zhao, L, Ke, B, et al.. Medmnist v2-a large-scale lightweight benchmark for 2D and 3D biomedical image classification. Sci Data 2023;10:41. https://doi.org/10.1038/s41597-022-01721-8.Search in Google Scholar PubMed PubMed Central
127. Wang, J, Yuille, AL. Semantic part segmentation using compositional model combining shape and appearance. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2015:1788–97 pp.10.1109/CVPR.2015.7298788Search in Google Scholar
128. Ravi, N, Reizenstein, J, Novotny, D, Gordon, T, Lo, W-Y, Johnson, J, et al.. Accelerating 3D deep learning with PyTorch3D. arXiv preprint arXiv:2007.08501; 2020. https://doi.org/10.1145/3415263.3419160.Search in Google Scholar
129. Khalid, N, Qayyum, A, Bilal, M, Al-Fuqaha, A, Qadir, J. Privacy-preserving artificial intelligence in healthcare: techniques and applications. Comput Biol Med 2023:106848. https://doi.org/10.1016/j.compbiomed.2023.106848.Search in Google Scholar PubMed
130. Schwarz, CG, Kremers, WK, Therneau, TM, Sharp, RR, Gunter, JL, Vemuri, P, et al.. Identification of anonymous MRI research participants with face-recognition software. N Engl J Med 2019;381:1684–6. https://doi.org/10.1056/nejmc1908881.Search in Google Scholar PubMed PubMed Central
131. Gießler, F, Thormann, M, Preim, B, Behme, D, Saalfeld, S. Facial feature removal for anonymization of neurological image data. Curr Dir Biomed Eng 2021;7:130–4. https://doi.org/10.1515/cdbme-2021-1028.Search in Google Scholar
132. McLaughlin, J, Fang, S, Huang, J, Robinson, L, Jacobson, S, Foroud, T, et al.. Interactive feature visualization and detection for 3d face classification. In: 9th IEEE international conference on cognitive informatics (ICCI’10). IEEE; 2010:160–7 pp.10.1109/COGINF.2010.5599748Search in Google Scholar
133. Suzuki, K, Nakano, H, Inoue, K, Nakajima, Y, Mizobuchi, S, Omori, M, et al.. Examination of new parameters for sex determination of mandible using Japanese computer tomography data. Dentomaxillofacial Radiol 2020;49:20190282. https://doi.org/10.1259/dmfr.20190282.Search in Google Scholar PubMed PubMed Central
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorial
- Change of publication model for Biomedical Engineering/Biomedizinische Technik
- Research Articles
- Mechano-responses of quadriceps muscles evoked by transcranial magnetic stimulation
- A type-2 fuzzy inference-based approach enables walking speed estimation that adapts to inter-individual gait patterns
- DeepCOVIDNet-CXR: deep learning strategies for identifying COVID-19 on enhanced chest X-rays
- Prediction of muscular-invasive bladder cancer using multi-view fusion self-distillation model based on 3D T2-Weighted images
- Evaluation of the RF depositions at 3T in routine clinical scans with respect to the SAR safety to improve efficiency of MRI utilization
- A software tool for fabricating phantoms mimicking human tissues with designated dielectric properties and frequency
- MedShapeNet – a large-scale dataset of 3D medical shapes for computer vision
- Concept and development of a telemedical supervision system for anesthesiology in operating rooms using the interoperable communication standard ISO/IEEE 11073 SDC
Articles in the same Issue
- Frontmatter
- Editorial
- Change of publication model for Biomedical Engineering/Biomedizinische Technik
- Research Articles
- Mechano-responses of quadriceps muscles evoked by transcranial magnetic stimulation
- A type-2 fuzzy inference-based approach enables walking speed estimation that adapts to inter-individual gait patterns
- DeepCOVIDNet-CXR: deep learning strategies for identifying COVID-19 on enhanced chest X-rays
- Prediction of muscular-invasive bladder cancer using multi-view fusion self-distillation model based on 3D T2-Weighted images
- Evaluation of the RF depositions at 3T in routine clinical scans with respect to the SAR safety to improve efficiency of MRI utilization
- A software tool for fabricating phantoms mimicking human tissues with designated dielectric properties and frequency
- MedShapeNet – a large-scale dataset of 3D medical shapes for computer vision
- Concept and development of a telemedical supervision system for anesthesiology in operating rooms using the interoperable communication standard ISO/IEEE 11073 SDC

