Abstract
Vertebrate heart formation is a spatiotemporally regulated morphogenic process that initiates with bilaterally symmetric cardiac primordial cells migrating toward the midline to form a linear heart tube. The heart tube then elongates and undergoes a series of looping morphogenesis, followed by expansions of regions that are destined to become primitive heart chambers. During the cardiac morphogenesis, cells derived from the first heart field contribute to the primary heart tube, and cells from the secondary heart field, cardiac neural crest, and pro-epicardial organ are added to the heart tube in a precise spatiotemporal manner. The coordinated addition of these cells and the accompanying endocardial cushion morphogenesis yield the atrial, ventricular, and valvular septa, resulting in the formation of a four-chambered heart. Perturbation of progenitor cells’ deployment and differentiation leads to a spectrum of congenital heart diseases. Two of the genes that were recently discovered to be involved in cardiac morphogenesis are Numb and Numblike. Numb, an intracellular adaptor protein, distinguishes sibling cell fates by its asymmetric distribution between the two daughter cells and its ability to inhibit Notch signaling. Numb regulates cardiac progenitor cell differentiation in Drosophila and controls heart tube laterality in Zebrafish. In mice, Numb and Numblike, the Numb family proteins (NFPs), function redundantly and have been shown to be essential for epicardial development, cardiac progenitor cell differentiation, outflow tract alignment, atrioventricular septum morphogenesis, myocardial trabeculation, and compaction. In this review, we will summarize the functions of NFPs in cardiac development and discuss potential mechanisms of NFPs in the regulation of cardiac development.
Introduction: numb and cardiac development
During the development of the Drosophila peripheral nervous system, a single sensory organ precursor (SOP) cell undergoes several divisions to produce four cells that form an external sensory organ (Figure 1A and B). In the first division of the SOP cell, Numb localizes asymmetrically at one pole of the mitotic cell cortex, so that only one daughter cell inherits the protein (Figure 1C). As a result, this daughter becomes a pIIb cell, and the other becomes a pIIa cell (Figure 1B and C) (1, 2). These two cells then divide to produce the different cell types of the sensory organ (Figure 1A and B). Numb gain or loss of function results in two IIb cell or IIa cells, respectively, and it was discovered that Numb promotes IIb cell fate by inhibiting Notch signaling (1, 3, 4).
Since then, many more functions of Numb have been revealed. It functions as a component of the adherens junctions to regulate cell adhesion and cell migration (5), and controls the stability of p53 (6) and Gli1 (7) to regulate cancer initiation. Numb has also been reported to complex with β-catenin and to regulate neuroepithelial and epicardial development (8, 9). The functions of Numb specifying neural cell fate are conserved in vertebrates (2, 4, 10–13).
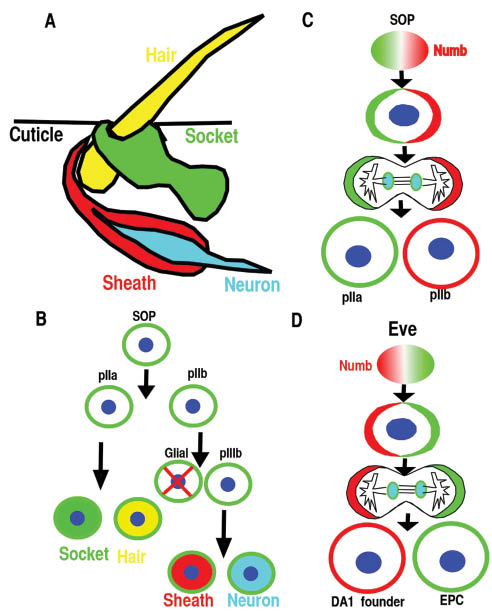
Numb is asymmetrically distributed during asymmetric cell division in different cell types.
(A, B) The Drosophila sensory organ consists of four cells: hair, socket, sheath, and neuron and is derived from the sensory organ precursor (SOP). (B, C) SOP divides asymmetrically in a stem cell-like fashion to generate the various cells of the sensory organ. The glial cell undergoes programmed cell death. (D) Eve-positive mesoderm progenitor cell divides asymmetrically to generate DA1 founder and eve-expressing pericardial cells (EPC).
Recently, Numb has been revealed to regulate cardiac progenitor cell differentiation and cardiac development in different species. In Drosophila, Numb is involved in specification of cardiac cell type via Notch signaling interference (14). In Zebrafish, Numb is required for heart left-right asymmetric morphogenesis via regulating Notch signaling (15). In mice, there are two homologs Numb and Numblike (16, 17). Numb is expressed in adult cardiac cKit cells and is asymmetrically distributed during their asymmetric cell divisions (18, 19). Furthermore, Numb and Numblike, the Numb family proteins (NFPs), are essential for cardiac morphogenesis and differentiation during development as evidenced by a variety of defects in cardiac morphogenesis and progenitor differentiation in the cardiac-specific NFP knockout embryos (20).
The vertebrates’ cardiac morphogenesis depends on the addition and differentiation of progenitor cells from four different sources (21) (Figure 2A–C). At approximately embryonic day 7.5 (E7.5), cardiac mesodermal cells arising in the anterior primitive streak migrate to the anterior ventral aspect to form a bilaterally symmetric heart field called the cardiac crescent (Figure 2A) (22, 23). The cardiac crescent, the source of the first two progenitor sources, consists of first heart field (FHF) and secondary heart field (SHF) with the SHF residing dorsomedially relative to FHF in the crescent (Figure 2A). Cells from FHF of the cardiac crescent will fold toward the ventral midline to form a linear heart tube at about E8.0 (Figure 2B). The SHF cells initially residing dorsomedially to FHF are subsequently located to the pharyngeal and splanchnic mesoderm, from which they migrate to the preexisting scaffold of the linear heart tube. The SHF cells will contribute to the right ventricle, OFT myocardium, and to some endocardium at E8.5–E10.25 (Figure 2C) (24–27). The cells derived from the SHF play an essential role in the orientation and patterning of the outflow tract (OFT) (28). Cardiac neural crest cells (CNCC), originating from postotic rhombomeres 6, 7, and 8, will migrate to the caudal pharynx and contribute significantly to the smooth muscle layer and endocardial cushion in the OFT (Figure 2C). They are also involved in the formation of the aorticopulmonary septum, as demonstrated by lineage-tracing studies using neural crest-restricted Cre mouse lines (29, 30). CNCCs are essential for normal myocardial differentiation in the OFT and for the formation and remodeling of the great arteries (31, 32). The fourth population is the epicardial cells derived from the pro-epicardial organ (PEO), which is located at the sinoatrial pole and atrioventricular junction at about E9.5 (Figure 2C). This population contributes to fibroblast, smooth muscle cell, and potentially other cardiac cell types (33). The epicardium regulates coronary vascular development, cardiac growth, and morphogenesis.
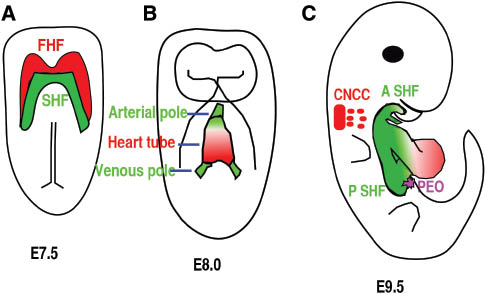
Four different cell sources contribute to heart formation.
(A) Ventral view of an E7.5 embryo. Red color highlights the cardiac progenitor cells of the first heart field (FHF), and green color highlights another subset of cardiac progenitor cells that form the secondary heart field (SHF) and is located posteriorly and medially to the FHF. (B) Ventral view of the embryo at E8.5. Cells from the FHF migrate and then merge in the midline to form the heart tube, which then elongates on both arterial and venous poles via the addition of progenitor cells from the SHF. (C) Right lateral view of an E9.5 mouse embryo. The cardiac neural crest cells (CNCC) at rhombomere 6–8 gives rise to cells (green) that migrate to and colonize the distal cardiac outflow tract (OFT) to contribute to OFT alignment and septation. Cells from pro-epicardial organ (PEO) will migrate, attach, and then cover the whole heart to form epicardium. A SHF, anterior second heart field; P SHF, posterior second heart field.
In a delicate structure like the heart, abnormal cell migration/differentiation during morphogenesis will cause malformations or congenital heart defects (CHD), which is the number one cause of birth defects in the world. Understanding how the heart is assembled at the cellular and molecular level is an essential step toward improving diagnosis and potential treatments for CHD. The variety of defects resulting from NFP deletions in different cardiac cell types indicates that NFPs are essential and novel factors involved in heart morphogenesis and progenitor differentiation.
NFPs function in cardiac cell specification and differentiation
Numb was identified as the first cell fate determinant in Drosophila (1) due to its ability to inhibit Notch signaling via endocytosis, and its functions are conserved during the specification of neural cell fate in mammals (2, 4, 10–13). Numb’s function as a cell fate determinant for cardiac cell was initially studied in Drosophila. Cardiogenesis in Drosophila can be considered as a series of distinct developmental decisions. These include the sequential specification of mesoderm, dorsal mesoderm, and cardiac fate within the dorsal mesoderm, followed by the cell fate diversification of the cardioblast in each segment and the cardiac cell types in the anterior-posterior heart tube (34). Numb mutant Drosophila is not defective in cardiac fate specification, but Numb is involved in the specification and differentiation of cardiac cell types such as pericardial cells at later stages of heart development. During the specification of pericardial cells, even skipped-expressing (Eve) myogenic progenitors divide asymmetrically, and Numb is asymmetrically distributed with one daughter cell inheriting the majority of Numb (Figure 1D). The presence of Numb inhibits Notch and Sanpodo signaling and causes this daughter cell to take the muscle founder cell fate (DA1). The other daughter cell that does not inherit Numb will take eve-expressing pericardial cell fate (Figure 1D). Disruption of Numb results in more pericardial cells, while overexpressing Numb reduces the number of pericardial cells and induces more DA1 cells (35, 36). When Sanpodo, which is required for Notch signaling, is disrupted, the mutant displayed an opposite phenotype to the Numb mutant (36). Within individual segments of Drosophila heart, there are two nonidentical groups of cardiac cells: four pairs of cardioblasts express Tinman (Tin) and two pairs of cardioblasts alternating with the Tin-expressing cells express Seven Up (Svp). The Tin-expressing cells are generated by symmetric cell divisions from cardiac progenitor cells (37). The Svp-expressing cells are generated by asymmetric cell divisions of heart progenitor cells in the mesoderm, with its sister cell becoming the Odd-pericardial cells (38). Numb promotes Svp-expressing cardiac cell at the expense of Odd-expressing pericardial cells. In the absence of Numb, Svp-expressing cells are not observed, while the number of Tin-expressing cells did not change (37), indicating that Numb is involved in cell specification only during asymmetric cell division (14).
In the mouse, NFPs global double knockout embryos die around E9.0 (12, 20). Whether the knockout displays a cardiac progenitor specification defect is not clear. The early embryonic lethality of global double knockout prevents the studying of NFP functions in later stages of cardiac development, but this can be overcome by conditional knockout technology. NFPs’ role in cardiac progenitor cell differentiation and renewal has been investigated with multiple Cre lines, which allow NFPs’ deletion at different stages. NFP deletion via the Mesp1-Cre, which is active in the mesoderm, disrupted cardiac progenitor renewal due to reduced proliferation, which results in hypoplastic OFT and right ventricle (39). The mechanism of how NFPs regulate cardiac progenitor self-renewal is not clear. NFPs regulate epicardial development and cardiac progenitor differentiation at later stages, and their disruptions at these stages cause various defects (9, 20, 40). In the following sections, we will discuss the functions of NFP in epicardial development, SHF progenitor cell differentiation, outflow tract morphogenesis, atrioventricular septation, and myocardial trabeculation.
NFPs are required for epicardial development by maintaining polarity
The epicardium, the outer cell layer of the heart, is composed of a single layer of epithelial cells that arises from the PEO (Figure 2C). Distinct compartments of PEO can be labeled by different molecular markers and contribute to different cardiac cell types (41). The pro-epicardial cells migrate from the PEO, attach, and then spread to cover the whole heart beginning at E9.5 (33, 42). Signaling pathways that regulate epicardial cell detachment from the PEO are not clear, while pathways that regulate epicardial cell attachment to the heart and subsequent entry into the myocardium have been extensively studied. α4β1 Integrin, which are expressed by epicardial cells (43), interact with fibronectin (44) and VCAM-1 (45–47) expressed by myocardial cells to promote epicardial cell adherence to cardiomyocytes and spreading to cover the heart (43, 48). After attaching to the heart, a subset of epicardial cells undergoes an epithelial-to-mesenchymal transition (EMT) and migrates into the myocardium (49–54). They subsequently differentiate into fibroblasts, smooth muscle cells, endocardial cells, and potentially cardiomyocytes (41, 55–63). Many signaling pathways, such as FGF (64, 65), PDGF (66), Wnt/β-catenin (51, 67), RXRα (50, 68, 69), TGFβ (54), and Notch (53, 70), play a role in these processes. Other proteins expressed by epicardial cells such as Par3 (71), GATA4 (72), WT1 (67, 73–75), α4β1 integrin, and podoplanin (76) are also required for proper epicardial development.
In epicardium, Numb localizes to the adherens junctions of epicardial cells at the G1 phase and to the basal domain at M phase (Figure 3A) (9), similar to its localization in neuroepithelium (8). Polarity proteins such as Par3, Par6, and aPKC localize to the apical domain of epicardial cells (9, 71), indicating that epicardial cells are polarized in a manner similar to other epithelial cells (77). NFPs are required to maintain epicardial polarity (9). Although multiple signaling pathways have been reported to regulate epicardial cell entry into the myocardium, the cellular mechanism is not clear. Time-lapse imaging and immunofluorescence staining enabled the discovery of epicardial cells that undergo parallel or perpendicular divisions with respect to the heart wall. The parallel divisions produce daughter cells to cover the heart, while a perpendicular division’s daughter cell invades the myocardium (Figure 3) (9). NFPs are required for β-catenin to localize to adherens junctions. Conditional deletion of NFPs in epicardium results in disruption of epicardial adherens junction, epicardial polarity, and random mitotic spindle orientations, which might cause the observed epicardial EMT defects (9). The epicardial cell perpendicular division is an asymmetric cell division, as the daughter cell that enters into the myocardium inherits more Numb and differentiates into a fibroblast or other cell types. The other daughter cell remaining within the epicardium maintains the epicardial cell fate. NFPs might play multiple roles in epicardial development. First, NFPs are required to stabilize adherens junctions, which are required to establish the mitotic spindle orientation. NFPs might regulate the stability of the components of adherens junctions such as β-catenin via endocytosis in epicardium. Second, Numb accumulation at the basal domain of the dividing epicardial cell beginning at the S-phase might also promote epicardial cell migration and differentiation. However, further experiments will be needed to determine whether and how NFPs regulate epicardial cell migration and differentiation.
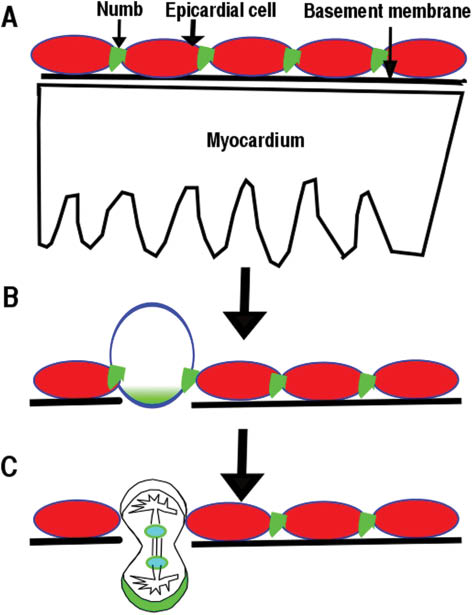
Epicardial cells’ perpendicular division contributes to their entry into myocardium.
(A) Numb localizes to adherens junctions of epicardial cells. (B) Numb accumulates at the basal domain of dividing epicardial cells. (C) One daughter cell from a perpendicular division will enter into the myocardium.
NFPs regulate second heart field progenitor cell differentiation
In 2001, three groups using various methods including viral infection tracing, vital dye lineage tracing, tissue ablation, and enhancer trap tracing demonstrated that cells in the splanchnic mesoderm migrated to the elongating cardiac linear tube and gave rise to the right ventricle and OFT. These observations led to the discovery of the SHF (78–80) and changed our view of cardiogenesis. We now know the FHF gives rise to the left ventricle and part of the inflow region, while the right ventricle, OFT, interventricular septum, endocardium, and part of the inflow region are derived from the SHF (26, 78–82).
Perturbation of SHF deployment and progenitor differentiation leads to a spectrum of CHDs. Several signaling pathways including FGF (78, 83, 84), Wnt (85–88), Hedgehog (89, 90), Notch (91), BMP (92), and retinoic acid (93) are involved in the deployment of Isl1 cells to the elongating linear heart tube and subsequent differentiation (24, 94, 95). FGF8 signaling functions upstream of Isl1, and deletion of Fgf8 specifically in SHF causes SHF morphogenesis defects (78, 83, 96). Type 1 BMP receptor deletion and BMP4 deletion decrease the proliferation in SHF and result in OFT septation defects (92, 97, 98). Hedgehog is crucial for cardiac neural crest cell survival and is also required for OFT septation (89, 99, 100).
NFPs deletion via Nkx2.5Cre/+ results in higher expression of progenitor markers such as Isl1, Tbx1, Fgf8, and Shox2 at E10.5. Isl1 expressional level in the knockout is significantly higher than that in the control littermates from E9.5 to E11.5. In addition, at E12.5 and E13.5, the knockouts displayed abnormal expressional levels of cardiomyocyte maturation/differentiation markers such as MYH6, MYH11, BMP10, Irx3-5, indicating a role for NFPs in cardiac progenitor differentiation. Supporting this notion, overexpressing Numb in the pluripotent stem cells in an embryoid body culture system promotes cardiac progenitor differentiation and decreases Isl1 expression in an endocytosis-dependent manner (20). Surprisingly, the differentiation defects in MDKO appear to be independent of Notch1, as Notch1 suppression in MDKO did not normalize the expression of these progenitor genes including Isl1. Instead, the upregulation of Fgf8 in the MDKO might be responsible for the upregulation of Isl1. Moreover, NFP regulation of Isl1 progenitor cells might be a stage or niche-dependent manner. Mesp1-Cre, which is active earlier than Nkx2.5Cre/+, mediated NFP deletion and reduced the number of Isl1 cells, possibly due to different niches (39).
NFP regulate outflow tract morphogenesis
The OFT is a single vascular conduit that links the right ventricle to the aortic sac. The septation of OFT into the aorta and pulmonary artery ensures blood flow from the right ventricle to the lung, back to the heart, and through the aorta to the whole body. Abnormal morphogenesis of the OFT, such as persistent truncus arteriosus (PTA), transposition of great arterial, double outlet right ventricle, and tetralogy of Fallot, causes shunting of oxygenated and deoxygenated blood. Understanding the regulatory pathways that control SHF deployment and progenitor differentiation in the OFT is essential to the understanding of the etiology of these CHDs.
The OFT is formed by several developmentally distinct cell populations, including cardiomyocytes derived from SHF, vascular smooth muscle cells from CNCC and SHF, and endothelial cells from SHF (21). Progenitor cells from SHF migrate to the OFT and differentiate into cardiomyocytes and smooth muscle cells at the arterial pole (101). The SHF-derived myocardium gives rise to the conotruncal myocardium, which is dependent on CNCC and is critical for normal alignment of the two arteries with respect to the ventricles (27, 102). This dependence is evident when CNCC ablation results in PTA, and failure of addition of SHF myocardium leads to malalignment of the arterial pole with the ventricles. The development of SHF and CNCCs are interdependent, as ablation of CNCCs results in changes in OFT length, where a loss of Fgf8 can affect both CNCC and SHF development (32, 83, 84, 102, 103).
NFP deletion via Nkx2.5Cre/+ results in OFT alignment defect, delayed OFT septation, and AVSD. Wnt1-Cre, which is active in CNCC, mediated NFPs deletion, does not cause any cardiac morphogenetic defect, indicating that NFPs in CNCC are not essential for cardiac morphogenesis and are not responsible for cardiac defects in MDKO. NFP deletion via Mef2c-Cre, which is active in SHF, recapitulates the morphogenetic defects in MDKO, indicating that NFPs in the SHF are essential for OFT alignment, OFT septation, and atrioventricular septal morphogenesis. Cardiomyocytes in the Mef2c-Cre-mediated NFP knockouts fail to form a myocardial spike, indicating a differentiation defect (20). NFP deletion with SM22-Cre, which is active in cardiomyocytes and smooth muscle cells at a later stage compared to Nkx2.5Cre/+, does not show defects in OFT alignment, OFT septation, or atrioventricular septal morphogenesis, which further support the notion that the morphogenesis defects in MDKO might be due to a cardiac progenitor cell differentiation defect in the SHF. SHF cells give rise to OFT myocardium and smooth muscle cells at the base of the aorta and pulmonary trunk to facilitate the separation of the aorta and pulmonary artery (26, 104). Whether NFPs are involved in the cell fate decision between cardiomyocytes and smooth muscle cells or biasing their fate during the OFT alignment/OFT septation is unknown.
NFP regulates atrioventricular septation via DMP formation
Atrioventricular septation is a complex morphogenetic process required for the formation of the four-chambered heart. There are five mesenchymal/muscular tissues involved in this atrioventricular septation: the superior and inferior atrioventricular endocardial cushions (AVC), the mesenchymal cap enveloped muscular atrial septum, the dorsal mesenchymal protrusion (DMP), and the interventricular muscular septum (Figure 4A and B) (105). The superior and inferior AVCs derive their mesenchymal cells from the endocardium through EMT. The continued growth of this mesenchyme ensures their fusion at the AV canal to separate atria from ventricles, and divides the AV canal into mitral and tricuspid orifices. DMP-derived cells proliferate and migrate through the dorsal mesocardium and bulge into the atrial chamber as a mesenchymal protrusion to reach other mesenchymal tissues (106). Simultaneously, the primary atrial septum grows from the atrial roof toward the AVC and merges with the AVC and DMP, anteriorly and posteriorly, respectively . The atrial muscular outgrowth partially septates the atrial chamber to the left and right atria. Within the ventricular chamber, an interventricular muscular septum emerges between the primitive left and right ventricles from the apex and grows superiorly to fuse with AVC, dividing the ventricular chamber into left and right ventricles. These mesenchymal tissues are later muscularized to form sturdy septum and eventually to septate the heart into a four-chambered functional heart. The abnormal atrioventricular morphogenesis results in a variety of congenital heart defects. The AVSD in MDKO is not caused by the functions of NFP in the mesenchymal tissues, as endocardial and endothelial cell-specific Tie2-Cre-mediated NFP knockouts do not display any abnormal heart morphogenesis. Instead, it might be caused by the abnormal differentiation and migration of posterior SHF progenitor cell.
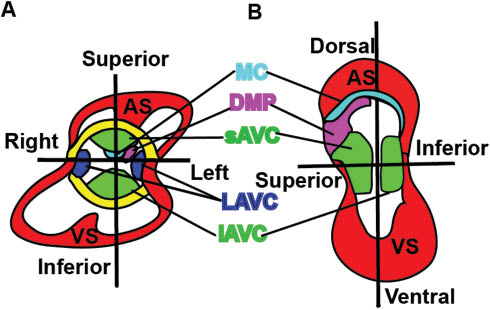
Five mesenchymal /muscular tissues contribute to atrioventricular septation.
(A and B) Different views of an E12–E13 heart. There are five mesenchymal /muscular tissues including the superior (sAVC) and inferior atrioventricular endocardial cushions (iAVC), lateral AV cushions (LAVC), the mesenchymal cap (MC) enveloped muscular atrial septum (AS), the dorsal mesenchymal protrusion (DMP), and the interventricular muscular septum (VS) that are involved in this atrioventricular septation (A and B).
The SHF gives rise to two spatially different populations: the anterior SHF, which is adjacent to the arterial pole, and the posterior SHF, which is adjacent to the venous pole (108). The posterior SHF contributes to the DMP, an essential structure for chamber septation (109). Abnormal differentiation and development of the posterior SHF has been associated with cardiac morphogenesis defects, such as atrial septal defect and atrioventricular septal defect (AVSD) (106, 110, 111). Nkx2.5Cre/+ and Mef2c-Cre-mediated NFP mutants lack the DMP, which is required for atrioventricular septation (112). Using lineage tracing and 3D imaging, we observed that cells derived from the SHF migrated to the AVC and formed the DMP in the control, but fewer or no cells were observed in the DMP in the knockout. NFPs have been reported to be involved in cell migration via recycling of different integrin subunits to the leading edge of migrating cells (113). This recycling might be involved in progenitor cell migration in the posterior SHF to form the DMP and the atrioventricular septum. Other possible mechanisms are either that NFPs regulate Isl1 cells to differentiate to the cardiac through Hedgehog signaling, etc., or NFPs maintain the cell-cell junctions via endocytosis to regulate the morphogenesis of DMP. However, more work will be needed to determine the detailed molecular mechanisms used by NFPs to regulate posterior SHF development.
NFP regulation of trabeculation and compaction extends beyond Notch 1 signaling inhibition
Trabeculae are sheet-like structures extending from the myocardium to the heart lumen (114). A lack of trabeculation causes embryonic lethality in mice, and excess trabeculation causes cardiomyopathy and heart failure in humans (115–117). Trabecular morphogenesis is a multiple-step process including, but not limited to, trabecular initiation, trabecular proliferation/growth, trabecular differentiation, and trabecular compaction. Trabeculation initiates at E9.0–E9.5 (118, 119). Trabecular cardiomyocytes proliferate at a similar rate as cardiomyocytes in the compact zone at an early stage, then gradually at a lower rate during later stages. Trabecular cardiomyocytes become more differentiated than cells in the compact zone. Myocardial compaction occurs at about E14.5, and the failure of trabeculae to coalesce with the compact zone is defined as left ventricular noncompaction (LVNC, OMIM300183) (120). Disruptions of genes that code components of the sarcomere and the Z-disk cause noncompaction (121, 122). Adding a layer of complexity is the physical environment, as the hemodynamics is required for trabeculation (123).
Signaling between endocardium and myocardium such as Brg1/ADAMTS1 (124), NRG1/ErBb2,4 (125–127), EphrinB2/EphB4 (128, 129), BMP10 (130), and Notch1 are required for trabeculation. Notch1 signaling activation in endocardium regulates trabeculation in an instructing manner (131). Global or endothelial-specific Notch1 deletion causes ventricular hypoplasia and trabeculation defects (131). The observation that both Notch1 loss (131) and gain of function (132) in endocardial cells reduces trabeculation indicates that mechanisms of Notch1 regulation of trabeculation are not clear.
The mechanism that regulates myocardium compaction is much less clear. One of the potential pathways is Notch2 signaling. Notch2 intracellular domain (N2ICD) is detected throughout the myocardium before E11.5, while at a later stage, Notch2 activity is specifically downregulated in the compact zone and is restricted to trabecular myocardium during ventricular compaction (40). This indicates that Notch2 might be involved in myocardial compaction. As further evidence, Notch2 global knockout displays ventricular hypoplasia (133), and its deletion in the heart via SM22-Cre results in cyanosis at birth due to narrowed arteries. Whether or not the knockout displayed a trabeculation defect was not reported (134).
Interestingly, MDKO displays defects in trabecular initiation, trabecular growth, differentiation, and compaction, indicating that NFPs are essential for trabeculation. MDKO hearts displays a higher level of Notch1 intracellular domain (N1ICD). N1ICD is detected in the myocardium of MDKO but not of the control. This suggests that NFPs inhibit N1ICD accumulation in cardiomyocytes. The upregulation of Notch signaling in MDKO is further confirmed with the transgenic mouse line that bears the canonical transgenic Notch reporter (TNR). This is the first in vivo evidence that NFPs inhibit Notch signaling in mammalian system. While all cardiomyocytes display upregulation of N1ICD, only some cardiomyocytes are TNR positive, indicating that NFP deletion upregulates both canonical and noncanonical Notch signaling. Genetic epistasis showed that Notch1 upregulation is responsible for decreased p57 expression and increased proliferative rate in trabeculae, and increased trabecular thickness. However, surprisingly, Notch1 suppression did not rescue the defects of trabecular initiation and noncompaction (20). Another target of NFPs is Notch2. In the control, N2ICD is only present in the trabeculae from E11.5 on, while N2ICD continues to be present in the compact zone of MDKO. N2ICD overexpression in cardiomyocytes mediated by αMHC-Cre results in hypertrabeculation and noncompaction, indicating that Notch2 is involved in compaction, and NFPs might inhibit Notch2 to regulate compaction (40). In summary, Notch1 and Notch2 might regulate different steps of trabecular morphogenesis. We speculate that NFPs inhibit Notch1 to regulate cardiomyocyte proliferation and trabecular growth/thickness, but not trabecular initiation and compaction, and that NFPs inhibit Notch2 to regulate compaction. However, many questions regarding the regulation of trabecular morphogenesis by NFPs remain. For instance, knowing how NFPs inhibit the proliferation and promote differentiation of trabecular cardiomyocytes and knowing how NFPs regulate trabecular initiation will yield a deeper understanding of the relationship between NFP and trabeculation. A broad understanding of how the signaling pathways control cellular dynamics during trabecular initiation and morphogenesis, particularly in the mammalian heart, remains to be clarified.
Conclusions and future questions
In summary, Numb is required to diversify cardiac cell types in Drosophila. In mice, NFPs play essential roles during cardiac development and cardiac progenitor cell differentiation. NFPs are required to establish epicardial polarity and epicardial cell mitotic spindle orientation to regulate epicardial cell entry into the myocardium. NFPs regulate cardiac progenitor cell proliferation early in development and differentiation at late stages. NFPs are required for outflow tract alignment and atrioventricular septation via controlling cardiac progenitor cell differentiation and migration. NFPs also inhibit Notch1 to regulate trabecular growth, and inhibit Notch2 to regulate myocardial compaction. Despite their essential functions during cardiac development, many questions still remain and prevent us from fully understanding how NFPs regulate cardiac development. The molecular mechanisms illuminating how NFPs regulate different biological processes via endocytosis are limited to the in vitro cultured system, and whether these molecular mechanisms also apply to the in vivo system is not clear. An endocytosis-defective Numb mouse model should be generated to answer this question. Also, NFPs interfere with signaling pathways at posttranslational level via endocytosis and recycling. The direct target of NFPs in different tissues may be distinct, which explains the various phenotypes in different tissue-specific knockouts. A high throughput screening such as quantitative proteomics will be needed to determine the direct targets in each tissue.
Acknowledgments
We thank members of the Wu Laboratory for discussions and advice. We thank Dr. John Schwarz for insight comments. We thank Ernest Spiotto and Thomas Myint for editing the manuscript. Sources of funding: work in the Wu Laboratory is supported by Albany Medical College start-up fund, the American Heart Association (13SDG16920099 to M.W.), and National Institute of Health (HL121700 to M.W.).
- List of abbreviations
- NFPs
Numb family proteins
- CHDs
congenital heart diseases
- SOP
sensory organ precursor
- FHF
first heart field
- SHF
secondary heart field
- OFT
outflow tract
- CNCC
cardiac neural crest cells
- PEO
pro-epicardial organ
- MDKO
Nkx2.5Cre/+-mediated myocardium Numb and Numblike double knockout
- AVSD
atrioventricular septal defect
- AVC
atrioventricular endocardial cushions
- DMP
dorsal mesenchymal protrusion
- EMT
epithelial-to-mesenchymal transition
- N1ICD
Notch1 intracellular domain
- N2ICD
Notch2 intracellular domain
- TNR
transgenic Notch reporter
References
1. Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 1994; 76: 477–91.10.1016/0092-8674(94)90112-0Search in Google Scholar
2. Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. Numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 1989; 58: 349–60.10.1016/0092-8674(89)90849-0Search in Google Scholar
3. Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of numb and notch. Neuron 1996; 17: 27–41.10.1016/S0896-6273(00)80278-0Search in Google Scholar
4. Spana EP, Doe CQ. Numb antagonizes notch signaling to specify sibling neuron cell fates. Neuron 1996; 17: 21–6.10.1016/S0896-6273(00)80277-9Search in Google Scholar
5. Wang Z, Sandiford S, Wu C, Li SS. Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. EMBO J 2009; 28: 2360–73.10.1038/emboj.2009.190Search in Google Scholar PubMed PubMed Central
6. Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP. NUMB controls p53 tumour suppressor activity. Nature 2008; 451: 76–80.10.1038/nature06412Search in Google Scholar PubMed
7. Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M, Screpanti I, Gulino A. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol 2006; 8: 1415–23.10.1038/ncb1510Search in Google Scholar PubMed
8. Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci 2007; 10: 819–27.10.1038/nn1924Search in Google Scholar PubMed
9. Wu M, Smith CL, Hall JA, Lee I, Luby-Phelps K, Tallquist MD. Epicardial spindle orientation controls cell entry into the myocardium. Dev Cell 2010; 19: 114–25.10.1016/j.devcel.2010.06.011Search in Google Scholar PubMed PubMed Central
10. Petersen PH, Zou K, Krauss S, Zhong W. Continuing role for mouse Numb and Numbl in maintaining progenitor cells during cortical neurogenesis. Nat Neurosci 2004; 7: 803–11.10.1038/nn1289Search in Google Scholar PubMed
11. Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, Temple S, Jan LY, Jan YN. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron 2003; 40: 1105–18.10.1016/S0896-6273(03)00755-4Search in Google Scholar
12. Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 2002; 419: 929–34.10.1038/nature01124Search in Google Scholar
13. Frise E, Knoblich JA, Younger-Shepherd S, Jan LY, Jan YN. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci USA 1996; 93: 11925–32.10.1073/pnas.93.21.11925Search in Google Scholar
14. Han Z, Bodmer R. Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development 2003; 130: 3039–51.10.1242/dev.00484Search in Google Scholar
15. Niikura Y, Tabata Y, Tajima A, Inoue I, Arai K, Watanabe S. Zebrafish Numb homologue: phylogenetic evolution and involvement in regulation of left-right asymmetry. Mech Dev 2006; 123: 407–14.10.1016/j.mod.2006.03.008Search in Google Scholar
16. Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron 1996; 17: 43–53.10.1016/S0896-6273(00)80279-2Search in Google Scholar
17. Verdi JM, Schmandt R, Bashirullah A, Jacob S, Salvino R, Craig CG, Program AE, Lipshitz HD, McGlade CJ. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr Biol 1996; 6: 1134–45.10.1016/S0960-9822(02)70680-5Search in Google Scholar
18. Cottage CT, Bailey B, Fischer KM, Avitable D, Collins B, Tuck S, Quijada P, Gude N, Alvarez R, Muraski J, Sussman MA. Cardiac progenitor cell cycling stimulated by pim-1 kinase. Circ Res 2010; 106: 891–901.10.1161/CIRCRESAHA.109.208629Search in Google Scholar PubMed PubMed Central
19. Wu M, Meng F. Has the cardiac stem cell controversy settled down? Sci China Life Sci 2014; 57: 949–50.10.1007/s11427-014-4690-6Search in Google Scholar PubMed
20. Zhao C, Guo H, Li J, Myint T, Pittman W, Yang L, Zhong W, Schwartz RJ, Schwarz JJ, Singer HA, Tallquist MD, Wu M. Numb family proteins are essential for cardiac morphogenesis and progenitor differentiation. Development 2014; 141: 281–95.10.1242/dev.093690Search in Google Scholar PubMed PubMed Central
21. Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol 2010; 90: 1–41.10.1016/S0070-2153(10)90001-XSearch in Google Scholar
22. Manasek FJ. Embryonic development of the heart. I. A light and electron microscopic study of myocardial development in the early chick embryo. J Morphol 1968; 125: 329–65.10.1002/jmor.1051250306Search in Google Scholar
23. Van Mierop LH. Embryology of the univentricular heart. Herz 1979; 4: 78–85.Search in Google Scholar
24. Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 2005; 6: 826–35.10.1038/nrg1710Search in Google Scholar
25. Kelly RG, Buckingham ME. The anterior heart – forming field: voyage to the arterial pole of the heart. Trends Genet 2002; 18: 210–6.10.1016/S0168-9525(02)02642-2Search in Google Scholar
26. Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol 2005; 287: 134–45.10.1016/j.ydbio.2005.08.041Search in Google Scholar PubMed
27. Ward C, Stadt H, Hutson M, Kirby ML. Ablation of the secondary heart field leads to tetralogy of Fallot and pulmonary atresia. Dev Biol 2005; 284: 72–83.10.1016/j.ydbio.2005.05.003Search in Google Scholar PubMed
28. Zhou W, Lin L, Majumdar A, Li X, Zhang X, Liu W, Etheridge L, Shi Y, Martin J, Van de Ven W, Kaartinen V, Wynshaw-Boris A, McMahon AP, Rosenfeld MG, Evans SM. Modulation of morphogenesis by noncanonical Wnt signaling requires ATF/CREB family-mediated transcriptional activation of TGFb2. Nat Genet 2007; 39: 1225–34.10.1038/ng2112Search in Google Scholar PubMed PubMed Central
29. Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development 1994; 120: 2213–24.10.1242/dev.120.8.2213Search in Google Scholar PubMed
30. Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development 2000; 127: 1607–16.10.1242/dev.127.8.1607Search in Google Scholar PubMed
31. Kirby ML, Gale TF, Stewart DE. Neural crest cells contribute to normal aorticopulmonary septation. Science 1983; 220: 1059–61.10.1126/science.6844926Search in Google Scholar PubMed
32. Waldo KL, Lo CW, Kirby ML. Connexin 43 expression reflects neural crest patterns during cardiovascular development. Dev Biol 1999; 208: 307–23.10.1006/dbio.1999.9219Search in Google Scholar
33. Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec 1981; 201: 157–68.10.1002/ar.1092010117Search in Google Scholar
34. Bryantsev AL, Cripps RM. Cardiac gene regulatory networks in Drosophila. Biochim Biophys Acta 2009; 1789: 343–53.10.1016/j.bbagrm.2008.09.002Search in Google Scholar
35. Carmena A, Murugasu-Oei B, Menon D, Jimenez F, Chia W. Inscuteable and numb mediate asymmetric muscle progenitor cell divisions during Drosophila myogenesis. Genes Dev 1998; 12: 304–15.10.1101/gad.12.3.304Search in Google Scholar
36. Park M, Yaich LE, Bodmer R. Mesodermal cell fate decisions in Drosophila are under the control of the lineage genes numb, Notch, and sanpodo. Mech Dev 1998; 75: 117–26.10.1016/S0925-4773(98)00098-7Search in Google Scholar
37. Gajewski K, Choi CY, Kim Y, Schulz RA. Genetically distinct cardial cells within the Drosophila heart. Genesis 2000; 28: 36–43.10.1002/1526-968X(200009)28:1<36::AID-GENE50>3.0.CO;2-4Search in Google Scholar
38. Ward EJ, Skeath JB. Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development 2000; 127: 4959–69.10.1242/dev.127.22.4959Search in Google Scholar
39. Shenje LT, Andersen P, Uosaki H, Fernandez L, Rainer PP, Cho GS, Lee DI, Zhong W, Harvey RP, Kass DA, Kwon C. Precardiac deletion of Numb and Numblike reveals renewal of cardiac progenitors. eLife (Cambridge) 2014; 3: e02164.10.7554/eLife.02164Search in Google Scholar
40. Yang J, Bucker S, Jungblut B, Bottger T, Cinnamon Y, Tchorz J, Müller M, Bettler B, Harvey R, Sun QY, Schneider A, Braun T. Inhibition of notch2 by numb/numblike controls myocardial compaction in the heart. Cardiovasc Res 2012; 96: 276–85.10.1093/cvr/cvs250Search in Google Scholar
41. Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, Tabin CJ. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell 2012; 22: 639–50.10.1016/j.devcel.2012.01.012Search in Google Scholar
42. Komiyama M, Ito K, Shimada Y. Origin and development of the epicardium in the mouse embryo. Anat Embryol (Berl) 1987; 176: 183–9.10.1007/BF00310051Search in Google Scholar
43. Yang JT, Rayburn H, Hynes RO. Cell adhesion events mediated by a4 integrins are essential in placental and cardiac development. Development 1995; 121: 549–60.10.1242/dev.121.2.549Search in Google Scholar
44. Orgogozo V, Schweisguth F, Bellaiche Y. Binary cell death decision regulated by unequal partitioning of Numb at mitosis. Development 2002; 129: 4677–84.10.1242/dev.129.20.4677Search in Google Scholar
45. Hession C, Osborn L, Goff D, Chi-Rosso G, Vassallo C, Pasek M, Pittack C, Tizard R, Goelz S, McCarthy K. Endothelial leukocyte adhesion molecule 1: direct expression cloning and functional interactions. Proc Natl Acad Sci USA 1990; 87: 1673–7.10.1073/pnas.87.5.1673Search in Google Scholar
46. Osborn L, Hession C, Tizard R, Vassallo C, Luhowskyj S, Chi-Rosso G, Lobb R. Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell 1989; 59: 1203–11.10.1016/0092-8674(89)90775-7Search in Google Scholar
47. Kwee L, Baldwin HS, Shen HM, Stewart CL, Buck C, Buck CA, Labow MA. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development 1995; 121: 489–503.10.1242/dev.121.2.489Search in Google Scholar
48. Sengbusch JK, He W, Pinco KA, Yang JT. Dual functions of a4b1 integrin in epicardial development: initial migration and long-term attachment. J Cell Biol 2002; 157: 873–82.10.1083/jcb.200203075Search in Google Scholar
49. Perez-Pomares JM, Macias D, Garcia-Garrido L, Munoz-Chapuli R. Contribution of the primitive epicardium to the subepicardial mesenchyme in hamster and chick embryos. Dev Dyn 1997; 210: 96–105.10.1002/(SICI)1097-0177(199710)210:2<96::AID-AJA3>3.0.CO;2-4Search in Google Scholar
50. Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Izpisua Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid X receptor a is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci USA 2005; 102: 18455–60.10.1073/pnas.0504343102Search in Google Scholar
51. Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci USA 2007; 104: 18109–14.10.1073/pnas.0702415104Search in Google Scholar
52. Huang X, Gao X, Diaz-Trelles R, Ruiz-Lozano P, Wang Z. Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev Biol 2008; 319: 258–66.10.1016/j.ydbio.2008.04.020Search in Google Scholar PubMed
53. Yang K, Doughman YQ, Karunamuni G, Gu S, Yang YC, Bader DM, Watanabe M. Expression of active Notch1 in avian coronary development. Dev Dyn 2009; 238: 162–70.10.1002/dvdy.21811Search in Google Scholar PubMed PubMed Central
54. Sridurongrit S, Larsson J, Schwartz R, Ruiz-Lozano P, Kaartinen V. Signaling via the Tgf-b type I receptor Alk5 in heart development. Dev Biol 2008; 322: 208–18.10.1016/j.ydbio.2008.07.038Search in Google Scholar PubMed PubMed Central
55. Wada AM, Smith TK, Osler ME, Reese DE, Bader DM. Epicardial/mesothelial cell line retains vasculogenic potential of embryonic epicardium. Circ Res 2003; 92: 525–31.10.1161/01.RES.0000060484.11032.0BSearch in Google Scholar PubMed
56. Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with in growth of the epicardial organ. Dev Biol 1996; 174: 221–32.10.1006/dbio.1996.0068Search in Google Scholar PubMed
57. Poelmann RE, Gittenberger-de Groot AC, Mentink MM, Bokenkamp R, Hogers B. Development of the cardiac coronary vascular endothelium, studied with antiendothelial antibodies, in chicken-quail chimeras. Circ Res. 1993; 73: 559–68.10.1161/01.RES.73.3.559Search in Google Scholar
58. Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 2006; 127: 607–19.10.1016/j.cell.2006.08.052Search in Google Scholar PubMed
59. Wessels A, Perez-Pomares JM. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat Rec A Discov Mol Cell Evol Biol 2004; 276: 43–57.10.1002/ar.a.10129Search in Google Scholar PubMed
60. Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR, Riley PR. Thymosin b4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007; 445: 177–82.10.1038/nature05383Search in Google Scholar PubMed
61. Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res 1998; 82: 1043–52.10.1161/01.RES.82.10.1043Search in Google Scholar
62. Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 2008; 454: 109–13.10.1038/nature07060Search in Google Scholar PubMed PubMed Central
63. Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature 2008; 454: 104–8.10.1038/nature06969Search in Google Scholar PubMed PubMed Central
64. Lavine KJ, Long F, Choi K, Smith C, Ornitz DM. Hedgehog signaling to distinct cell types differentially regulates coronary artery and vein development. Development 2008; 135: 3161–71.10.1242/dev.019919Search in Google Scholar PubMed PubMed Central
65. Morabito CJ, Dettman RW, Kattan J, Collier JM, Bristow J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev Biol 2001; 234: 204–15.10.1006/dbio.2001.0254Search in Google Scholar PubMed
66. Mellgren AM, Smith CL, Olsen GS, Eskiocak B, Zhou B, Kazi MN, Ruiz FR, Pu WT, Tallquist MD. Platelet-derived growth factor receptor b signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ Res 2008; 103: 1393–401.10.1161/CIRCRESAHA.108.176768Search in Google Scholar PubMed PubMed Central
67. von Gise A, Zhou B, Honor LB, Ma Q, Petryk A, Pu WT. WT1 regulates epicardial epithelial to mesenchymal transition through b-catenin and retinoic acid signaling pathways. Dev Biol 2011; 356: 421–31.10.1016/j.ydbio.2011.05.668Search in Google Scholar PubMed PubMed Central
68. Chen TH, Chang TC, Kang JO, Choudhary B, Makita T, Tran CM, Burch JB, Eid H, Sucov HM. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev Biol 2002; 250: 198–207.10.1006/dbio.2002.0796Search in Google Scholar PubMed
69. Sucov HM, Dyson E, Gumeringer CL, Price J, Chien KR, Evans RM. RXR a mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev 1994; 8: 1007–18.10.1101/gad.8.9.1007Search in Google Scholar PubMed
70. del Monte G, Casanova JC, Guadix JA, MacGrogan D, Burch JB, Perez-Pomares JM, de la Pompa JL. Differential Notch signaling in the epicardium is required for cardiac inflow development and coronary vessel morphogenesis. Circ Res 2011; 108: 824–36.10.1161/CIRCRESAHA.110.229062Search in Google Scholar PubMed
71. Hirose T, Karasawa M, Sugitani Y, Fujisawa M, Akimoto K, Ohno S, Noda T. PAR3 is essential for cyst-mediated epicardial development by establishing apical cortical domains. Development 2006; 133: 1389–98.10.1242/dev.02294Search in Google Scholar PubMed
72. Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci USA 2004; 101: 12573–8.10.1073/pnas.0400752101Search in Google Scholar
73. Shao Y, Lu J, Zhang G, Liu C, Huang B. Histone acetyltransferase p300 promotes the activation of human WT1 promoter and intronic enhancer. Arch Biochem Biophys 2005; 436: 62–8.10.1016/j.abb.2005.01.007Search in Google Scholar
74. Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 1999; 126: 1845–57.10.1242/dev.126.9.1845Search in Google Scholar
75. Martinez-Estrada OM, Lettice LA, Essafi A, Guadix JA, Slight J, Velecela V, Hall E, Reichmann J, Devenney PS, Hohenstein P, Hosen N, Hill RE, Muñoz-Chapuli R, Hastie ND. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet 2010; 42: 89–93.10.1038/ng.494Search in Google Scholar
76. Mahtab EA, Wijffels MC, Van Den Akker NM, Hahurij ND, Lie-Venema H, Wisse LJ, Deruiter MC, Uhrin P, Zaujec J, Binder BR, Schalij MJ, Poelmann RE, Gittenberger-De Groot AC. Cardiac malformations and myocardial abnormalities in podoplanin knockout mouse embryos: correlation with abnormal epicardial development. Dev Dyn 2008; 237: 847–57.10.1002/dvdy.21463Search in Google Scholar
77. Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 2005; 437: 275–80.10.1038/nature03922Search in Google Scholar
78. Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell 2001; 1: 435–40.10.1016/S1534-5807(01)00040-5Search in Google Scholar
79. Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development 2001; 128: 3179–88.10.1242/dev.128.16.3179Search in Google Scholar PubMed
80. Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol 2001; 238: 97–109.10.1006/dbio.2001.0409Search in Google Scholar PubMed
81. Zaffran S, Kelly RG, Meilhac SM, Buckingham ME, Brown NA. Right ventricular myocardium derives from the anterior heart field. Circ Res 2004; 95: 261–8.10.1161/01.RES.0000136815.73623.BESearch in Google Scholar PubMed
82. Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell 2004; 6: 685–98.10.1016/S1534-5807(04)00133-9Search in Google Scholar
83. Ilagan R, Abu-Issa R, Brown D, Yang YP, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Fgf8 is required for anterior heart field development. Development 2006;133:2435–45.10.1242/dev.02408Search in Google Scholar PubMed
84. Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development 2006; 133: 2419–33.10.1242/dev.02367Search in Google Scholar PubMed PubMed Central
85. Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE. Biphasic role for Wnt/b-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci USA 2007; 104: 9685–90.10.1073/pnas.0702859104Search in Google Scholar PubMed PubMed Central
86. Kwon C, Arnold J, Hsiao EC, Taketo MM, Conklin BR, Srivastava D. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc Natl Acad Sci USA 2007; 104: 10894–9.10.1073/pnas.0704044104Search in Google Scholar PubMed PubMed Central
87. Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini A, Klein WH, Martin JF. Canonical Wnt signaling functions in second heart field to promote right ventricular growth. Proc Natl Acad Sci USA 2007; 104: 9319–24.10.1073/pnas.0701212104Search in Google Scholar PubMed PubMed Central
88. Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, Rosenfeld MG, Chen J, Evans SM. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci USA 2007; 104: 9313–8.10.1073/pnas.0700923104Search in Google Scholar PubMed PubMed Central
89. Washington Smoak I, Byrd NA, Abu-Issa R, Goddeeris MM, Anderson R, Morris J, Yamamura K, Klingensmith J, Meyers EN. Sonic hedgehog is required for cardiac outflow tract and neural crest cell development. Dev Biol 2005; 283: 357–72.10.1016/j.ydbio.2005.04.029Search in Google Scholar PubMed
90. Dyer LA, Kirby ML. Sonic hedgehog maintains proliferation in secondary heart field progenitors and is required for normal arterial pole formation. Dev Biol 2009; 330: 305–17.10.1016/j.ydbio.2009.03.028Search in Google Scholar PubMed PubMed Central
91. High FA, Jain R, Stoller JZ, Antonucci NB, Lu MM, Loomes KM, Kaestner KH, Pear WS, Epstein JA. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. J Clin Invest 2009; 119: 1986–96.10.1172/JCI38922Search in Google Scholar PubMed PubMed Central
92. Liu W, Selever J, Wang D, Lu MF, Moses KA, Schwartz RJ, Martin JF. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc Natl Acad Sci USA 2004; 101: 4489–94.10.1073/pnas.0308466101Search in Google Scholar PubMed PubMed Central
93. Ryckebusch L, Wang Z, Bertrand N, Lin SC, Chi X, Schwartz R, Zaffran S, Niederreither K. Retinoic acid deficiency alters second heart field formation. Proc Natl Acad Sci USA 2008; 105: 2913–8.10.1073/pnas.0712344105Search in Google Scholar PubMed PubMed Central
94. Black BL. Transcriptional pathways in second heart field development. Semin Cell Dev Biol 2007; 18: 67–76.10.1016/j.semcdb.2007.01.001Search in Google Scholar PubMed PubMed Central
95. Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res 2009; 104: 933–42.10.1161/CIRCRESAHA.109.194464Search in Google Scholar PubMed
96. Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development 2008; 135: 3599–610.10.1242/dev.025437Search in Google Scholar PubMed PubMed Central
97. Klaus A, Saga Y, Taketo MM, Tzahor E, Birchmeier W. Distinct roles of Wnt/b-catenin and Bmp signaling during early cardiogenesis. Proc Natl Acad Sci USA 2007; 104: 18531–6.10.1073/pnas.0703113104Search in Google Scholar PubMed PubMed Central
98. Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development 2006; 133: 1575–85.10.1242/dev.02322Search in Google Scholar PubMed PubMed Central
99. Lin L, Bu L, Cai CL, Zhang X, Evans S. Isl1 is upstream of sonic hedgehog in a pathway required for cardiac morphogenesis. Dev Biol 2006; 295: 756–63.10.1016/j.ydbio.2006.03.053Search in Google Scholar PubMed
100. Goddeeris MM, Schwartz R, Klingensmith J, Meyers EN. Independent requirements for Hedgehog signaling by both the anterior heart field and neural crest cells for outflow tract development. Development 2007; 134: 1593–604.10.1242/dev.02824Search in Google Scholar PubMed
101. Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, Abu-Issa R, Kirby ML. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev Biol 2005; 281: 78–90.10.1016/j.ydbio.2005.02.012Search in Google Scholar PubMed
102. Yelbuz TM, Waldo KL, Kumiski DH, Stadt HA, Wolfe RR, Leatherbury L, Kirby ML. Shortened outflow tract leads to altered cardiac looping after neural crest ablation. Circulation 2002; 106: 504–10.10.1161/01.CIR.0000023044.44974.8ASearch in Google Scholar
103. Hutson MR, Kirby ML. Neural crest and cardiovascular development: a 20-year perspective. Birth defects research Part C, Embryo Today Reviews 2003; 69: 2–13.10.1002/bdrc.10002Search in Google Scholar
104. Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell 2003; 5: 877–89.10.1016/S1534-5807(03)00363-0Search in Google Scholar
105. Lin CJ, Lin CY, Chen CH, Zhou B, Chang CP. Partitioning the heart: mechanisms of cardiac septation and valve development. Development 2012; 139: 3277–99.10.1242/dev.063495Search in Google Scholar PubMed PubMed Central
106. Snarr BS, Wirrig EE, Phelps AL, Trusk TC, Wessels A. A spatiotemporal evaluation of the contribution of the dorsal mesenchymal protrusion to cardiac development. Dev Dyn 2007; 236: 1287–94.10.1002/dvdy.21074Search in Google Scholar PubMed PubMed Central
107. Mommersteeg MT, Soufan AT, de Lange FJ, van den Hoff MJ, Anderson RH, Christoffels VM, Moorman AF. Two distinct pools of mesenchyme contribute to the development of the atrial septum. Cir Res 2006; 99: 351–3.10.1161/01.RES.0000238360.33284.a0Search in Google Scholar PubMed
108. Francou A, Saint-Michel E, Mesbah K, Theveniau-Ruissy M, Rana MS, Christoffels VM, Kelly RG. Second heart field cardiac progenitor cells in the early mouse embryo. Biochim Biophys Acta 2013; 1833: 795–8.10.1016/j.bbamcr.2012.10.003Search in Google Scholar PubMed
109. Snarr BS, O’Neal JL, Chintalapudi MR, Wirrig EE, Phelps AL, Kubalak SW, Wessels A. Isl1 expression at the venous pole identifies a novel role for the second heart field in cardiac development. Circ Res 2007; 101: 971–4.10.1161/CIRCRESAHA.107.162206Search in Google Scholar PubMed
110. Briggs LE, Kakarla J, Wessels A. The pathogenesis of atrial and atrioventricular septal defects with special emphasis on the role of the dorsal mesenchymal protrusion. Differentiation 2012; 84: 117–30.10.1016/j.diff.2012.05.006Search in Google Scholar PubMed PubMed Central
111. Hoffmann AD, Peterson MA, Friedland-Little JM, Anderson SA, Moskowitz IP. sonic hedgehog is required in pulmonary endoderm for atrial septation. Development 2009; 136: 1761–70.10.1242/dev.034157Search in Google Scholar PubMed PubMed Central
112. Wan J, Zhao S, Cheng H, Lu M, Jiang S, Yin G, Gao X, Yang Y. Varied distributions of late gadolinium enhancement found among patients meeting cardiovascular magnetic resonance criteria for isolated left ventricular non-compaction. J Cardiovasc Magn Reson 2013; 15: 20.10.1186/1532-429X-15-20Search in Google Scholar PubMed PubMed Central
113. Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell 2007; 13: 15–28.10.1016/j.devcel.2007.05.003Search in Google Scholar PubMed
114. Sedmera D, Thomas PS. Trabeculation in the embryonic heart. Bioessays 1996; 18: 607.10.1002/bies.950180714Search in Google Scholar PubMed
115. Jenni R, Rojas J, Oechslin E. Isolated noncompaction of the myocardium. N Engl J Med 1999; 340: 966–7.10.1056/NEJM199903253401215Search in Google Scholar PubMed
116. Breckenridge RA, Anderson RH, Elliott PM. Isolated left ventricular non-compaction: the case for abnormal myocardial development. Cardiol Young 2007; 17: 124–9.10.1017/S1047951107000273Search in Google Scholar PubMed
117. Weiford BC, Subbarao VD, Mulhern KM. Noncompaction of the ventricular myocardium. Circulation 2004; 109: 2965–71.10.1161/01.CIR.0000132478.60674.D0Search in Google Scholar PubMed
118. Zhang W, Chen H, Qu X, Chang CP, Shou W. Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC). Am J Med Genet C Semin Med Genet 2013; 163: 144–56.10.1002/ajmg.c.31369Search in Google Scholar PubMed PubMed Central
119. Samsa LA, Yang B, Liu J. Embryonic cardiac chamber maturation: trabeculation, conduction, and cardiomyocyte proliferation. Am J Med Genet C Semin Med Genet 2013; 163: 157–68.10.1002/ajmg.c.31366Search in Google Scholar PubMed PubMed Central
120. Pignatelli RH, McMahon CJ, Dreyer WJ, Denfield SW, Price J, Belmont JW, Craigen WJ, Wu J, El Said H, Bezold LI, Clunie S, Fernbach S, Bowles NE, Towbin JA. Clinical characterization of left ventricular noncompaction in children: a relatively common form of cardiomyopathy. Circulation 2003; 108: 2672–8.10.1161/01.CIR.0000100664.10777.B8Search in Google Scholar PubMed
121. Teekakirikul P, Kelly MA, Rehm HL, Lakdawala NK, Funke BH. Inherited cardiomyopathies: molecular genetics and clinical genetic testing in the postgenomic era. J Mol Diagn 2013; 15: 158–70.10.1016/j.jmoldx.2012.09.002Search in Google Scholar PubMed
122. Finsterer J. Cardiogenetics, neurogenetics, and pathogenetics of left ventricular hypertrabeculation/noncompaction. Pediatr Cardiol 2009; 30: 659–81.10.1007/s00246-008-9359-0Search in Google Scholar
123. Peshkovsky C, Totong R, Yelon D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev Dyn 2011; 240: 446–56.10.1002/dvdy.22526Search in Google Scholar
124. Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, Chang CP. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell 2008; 14: 298–311.10.1016/j.devcel.2007.11.018Search in Google Scholar
125. Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 1995; 378: 394–8.10.1038/378394a0Search in Google Scholar
126. Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature 1995; 378: 386–90.10.1038/378386a0Search in Google Scholar
127. Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 1995; 378: 390–4.10.1038/378390a0Search in Google Scholar
128. Gerety SS, Wang HU, Chen ZF, Anderson DJ. Symmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol Cell 1999; 4: 403–14.10.1016/S1097-2765(00)80342-1Search in Google Scholar
129. Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell 1998; 93: 741–53.10.1016/S0092-8674(00)81436-1Search in Google Scholar
130. Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, Chen Z, Yang Z, Schneider MD, Chien KR, Conway SJ, Yoder MC, Haneline LS, Franco D, Shou W. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 2004; 131: 2219–31.10.1242/dev.01094Search in Google Scholar PubMed PubMed Central
131. Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, Shou W, Ballestar E, Esteller M, Rojas A, Pérez-Pomares JM, de la Pompa JL. Notch signaling is essential for ventricular chamber development. Dev Cell 2007; 12: 415–29.10.1016/j.devcel.2006.12.011Search in Google Scholar PubMed PubMed Central
132. Venkatesh DA, Park KS, Harrington A, Miceli-Libby L, Yoon JK, Liaw L. Cardiovascular and hematopoietic defects associated with Notch1 activation in embryonic Tie2-expressing populations. Circ Res 2008; 103: 423–31. PubMed PMID: 18617694.10.1161/CIRCRESAHA.108.177808Search in Google Scholar PubMed PubMed Central
133. McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 2001; 128: 491–502.10.1242/dev.128.4.491Search in Google Scholar PubMed
134. Varadkar P, Kraman M, Despres D, Ma G, Lozier J, McCright B. Notch2 is required for the proliferation of cardiac neural crest-derived smooth muscle cells. Dev Dyn 2008; 237: 1144–52.10.1002/dvdy.21502Search in Google Scholar PubMed
©2015 by De Gruyter
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Articles in the same Issue
- Frontmatter
- Reviews
- Transgenerational epigenetic inheritance: resolving uncertainty and evolving biology
- Longevity: epigenetic and biomolecular aspects
- Stepping inside the realm of epigenetic modifiers
- Numb family proteins: novel players in cardiac morphogenesis and cardiac progenitor cell differentiation
- Short Conceptual Overviews
- Suggested roles for microRNA in tumors
- The TATA-box motif and its impact on transcriptional gene regulation by miRNAs
Articles in the same Issue
- Frontmatter
- Reviews
- Transgenerational epigenetic inheritance: resolving uncertainty and evolving biology
- Longevity: epigenetic and biomolecular aspects
- Stepping inside the realm of epigenetic modifiers
- Numb family proteins: novel players in cardiac morphogenesis and cardiac progenitor cell differentiation
- Short Conceptual Overviews
- Suggested roles for microRNA in tumors
- The TATA-box motif and its impact on transcriptional gene regulation by miRNAs

