Technical and health governance aspects of the External Quality Assessment Scheme for the SARS-CoV-2 molecular tests: institutional experience performed in all clinical laboratories of a Regional Health Service
Abstract
Objectives
Since December 2019, the worldwide public health has been threatened by a severe acute respiratory syndrome caused by Coronavirus-2. From the beginning, a turning point has been the identification of new cases of infection, in order to minimize the virus spreading among the population. For this reason, it was necessary introducing a panel of tests able to identify positive cases, which became crucial for all countries.
Methods
As a Regional Reference Centre, the CRQ Laboratory (Regional Laboratory for the Quality Control) developed and conducted an External Quality Assessment (EQA) panel of assay, so as to evaluate the quality of real-time reverse transcription polymerase chain reaction (PCR), which were used by 62 Sicilian laboratories, previously authorized to issue certificates for the COVID-19 diagnosis, on behalf of the Public Health Service.
Results
The qualitative performance test was based on pooled samples with different viral loads of SARS-CoV-2 or human Coronavirus OC43. 75% of the participating laboratories tested all core samples correctly, while the remaining 25% interpreted incorrectly the EQA exercise samples matching negatively the standards required.
Conclusions
Subsequent inspection visits confirmed the issue of incorrect positive and negative certifications for COVID-19 by private and public laboratories, despite the possession of the authorization requirements currently provided for by current regulations, with a significant impact on the SSR.
Introduction
As for June 20, 2022, the health emergency for the ongoing pandemic by Coronavirus infectious disease (COVID)-19, that caused a severe acute respiratory syndrome by Coronavirus-2 (SARS-CoV-2), has affected over 533 million people worldwide, resulting in over 6.3 million deaths [1]. Italy was the first European country fighting on the front lines of COVID-19, and a forerunner for the clinical and social interventions. On 20th of June 2022, the pandemic, in Italy, led to 17,773,764 confirmed cases and 167,617 deaths [2].
Typical clinical symptoms of most patients are fever, dry cough, hypoxemia and respiratory failure, typical of the acute respiratory distress syndrome (ARDS) [3].
The first viral genome sequence was released for immediate public health support via the community online resource virological.org on 10 January, 2020 (Wuhan-Hu-1, GenBank accession number MN908947) [4]. SARS-CoV-2 is a single stranded positive sense RNA virus, which shares 79% genome sequence identity with SARS-CoV and 50% with MERS-CoV [5]. The virus genome consists of six major open-reading frames (ORFs), arranged in the order of 5′ to 3′ as follows: replicase ORF1ab, spike (S), envelope (E), membrane (M) and nucleocapsid (N) [6].
In order to facilitate public health interventions with the aim of interrupting the transmission chain, early and accurate detection of SARS-CoV-2 with an efficient laboratory diagnosis becomes necessary. The WHO provided interim guidance for laboratory testing for COVID-2019 [7]. Specifically, real-time reverse transcription polymerase chain reaction (rRT-PCR) is routinely used to detect causative to diseases viruses from nasal and pharyngeal swabs, bronchoalveolar lavage, and blood plasma. The viral genes targeted include the N, E, S and RdRP genes [7]. Therefore, rRT-PCR needs to be continuously adapted to precisely identify new variants of concern (VOC) or seroptypes [8].
However, rRT-PCR tests show several technical and generic limitations. Firstly, rRT-PCR tests do not discriminate the results into a binary classification, either positive or negative, but in a semi-quantitative determination, based on cycle threshold (Ct) values and referencing to the viral load [9]. Secondly, rRT-PCR tests require 2–3 h to generate results, which are technically intricate and labor-intensive, that have higher costs, easily contaminated, and results being inefficient or wrong in case of virus mutation based on the assay used. Consequently, the standardization in reporting rRT-PCR Ct values it is necessary to assess the reliability of the laboratories tests providing the SARS-CoV-2 results as well as medical important decisions [10], [11], [12]. For these reasons, an External Quality Assessment (EQA) scheme, that include samples with known specific properties, allows to verify the skills of the participant laboratories. Therefore, the Regional Health Services authorized the execution of molecular biology investigations of SARS-Cov-2 for only specific and performing laboratories.
As an Institutional Public Provider for EQA, we developed and conducted an EQA in 62 laboratories, accredited and authorized by the Regional Health Service, in order to challenge the effectiveness of the laboratory’s quality management system for SARS-CoV-2 detection. In this report, we present the outcome of the EQA conducted in Sicily, and small surrounding islands (first in Italy) providing an overview of the diagnostic laboratory’s performances.
Materials and methods
EQA design
The Sicilian EQA system provides a mandatory participation in the SARS-CoV-2 EQA for every public and private laboratory that intends performing SARS-CoV-2 molecular testing to patients. The number of participating laboratories has increased sharply since the beginning of the pandemic, and thus has given more strength to the scientific data in our study.
Out of 654 laboratories belonging to the regional health service only 62 were enrolled for the participation in the EQA scheme “Molecular Biology SARS-COV-2 – COVID437” (340). These laboratories were the ones authorized to perform such analysis for the Regional Health System and for COVID-19 case identification. The authorization was released from the Regional Government Office after a specific and public selection based on a particular, structural, technological and professional features follow: (1) the presence of a molecular biologist; (2) the presence of a geneticist medical doctor or microbiologist; (3) the presence of DNA/RNA automatic extraction platforms; (4) the presence of validated Real-Time PCR systems.
Participants
The results of the participants were submitted through the CRQ’s EQA Platform. By the use of the CRQ Platform, the laboratory can manage the entire EQA process, from laboratory personal data registration and EQA orders, to the evaluation of EQA results, statistical analysis and reporting. As soon as a laboratory has been registered on the Platform, it can access to the CRQ EQA, which offers an on-line catalogue and submits its own EQA orders in an e-commerce manner. The catalogue is mainly structured into various categories, each of which give full details of EQA. The details include specimen type, sample quantity, analyte list, etc. Information about the scheduling of samples shipment and related quality test deadlines are provided, as well. A laboratory is updated about the status of its orders and samples shipments, so it has to confirm samples delivery on-line before it can access EQAs results submitting process. As a first step of SARS-CoV-2 EQA, a laboratory has to specify for each tested gene of the virus, instruments, reagents and methods used for every analysis phase: extraction, amplification and detection. Instruments, reagents and methods are not entered manually, but selected by accessing the Platform catalogues. The latter are constantly updated by the CRQ, and at the request of the laboratory. Before a laboratory enters analysis results, it has to specify if the conditions of the received samples are valid, so that the CRQ can take it into consideration for the evaluation phase. Subsequently, laboratories must enter the results of the analysis for each tested gene, expressed also in terms of CT values, including a medical report with the overall results of each samples (i.e. positive or negative for COVID-19). As a proof of the analysis performed, laboratories need to upload the reports produced by the instruments used, from which the entered results can be deduced.
Preparation of simulated samples
Briefly, 7 EQA samples (SARS-COV-2 Wuhan wild type) were shipped to each participant laboratory. In order to fulfill the quality criteria and to overcome health and legal issues, the 7 test samples were subministrated to the laboratories in a blind and name-inverted way, given that each scheme could reflect the real conditions of the patient. Coronavirus positive samples deployed in the Extra EQA scheme “Molecular Biology SARS-COV-2 – COVID437” derive from lysates of cells which have been infected with coronavirus (SARS-CoV-2, HCoV OC43 or HCoV 229E) and contain heat inactivated virus. In particular, for the EQA1 round, the positive samples represent different dilution series of lysate of cells infected with SARS-CoV-2, (1:1,000; 1:1,000,000; 1:10,000; 1:100,000). Negative samples derive from (1) lysates of non-infected cells MRC-5 as specificity control; (2) lysates of HCoV OC43-infected cells (1:2,500) as specificity control; and (3) lysate of HCoV 229E-infected cells (1:2,500) as specificity control. For the EQA2 round the positive samples were dilution series of lysate of SARS-CoV-2 infected cells (1:50,000,000; 1:5,000,000; 1:500,000; 1:50,000). Negative samples derive from (1) lysates of non-infected cells MRC-5 as specificity control; (2) lysates of HCoV OC43-infected cells lysates (1:1,000) as specificity control; and (3) lysate of HCoV NL63-infected cells (1:1,000) as specificity control. For the EQA3 round the positive samples were dilution series of lysate of SARS-CoV-2 infected cells (1:75,000; 1:50,000; 1:7,500; 1:5,000). Negative samples derive from (1) lysates of non-infected cells MRC-5 as specificity control; (2) lysates of HCoV OC43-infected cells lysates (1:1,000) as specificity control; and (3) lysate of HCoV 229-E-infected cells (1:1,000) as specificity control.
Each panel bearing the COVID437 code consists of 7 samples labeled A – G and consist of cell suspensions negative or positive for SARS-CoV-2 (genome complete viral) inactivated by heat. Each aliquot volume of approximately 0.8 mL is sufficient for at least two uses in normal nucleic acid extraction procedures.
Before the distribution to the participating laboratories of the EQA scheme, the target value of a given EQA sample is also confirmed in three ways: (1) technical support structure (Quality Control and Chemical and Biological Risk – CQRC) of the CRQ (Quality Control and Chemical) provider through two different PCR-RT assay systems: TaqPath COVID-19 CE-IVD RT-PCR (Applied Biosystems, Waltham, MA, USA) and Diagnostic kit for SARS-CoV-2 Nucleic Acid (Real-time PCR) (Shanghai Kehua bio-engineering Co., Ltd, Xuhui District, Shanghai, China); (2) INSTAND Expert Laboratories; (3) through the outcomes of the EQA scheme “Virus Genome Detection – SARS-CoV-2” (340) of the INSTAND, provider ISO/IEC 17043 accredited [13].
The target value includes median from Ct/Cp/Cq/CN values, lowest reported Ct/Cp/Cq/CN value and highest reported Ct/Cp/Cq/CN value for each gene analyzed (E, N, Orf1a, Orf1ab, RdRp, S, and a not specified region). Other samples are used for the detection of SARS-CoV-2 by PCR.
Data collection and statistical analysis
Statistical analysis, evaluation of the participants and reporting are performed by the CRQ’s EQA Platform engine. The evaluation takes into account the overall entered result of each sample against target results. Warnings about received samples, if any, are considered in this phase. Furthermore, uploaded instrument reports are evaluated. Participants achieve a positive evaluation if the target results are fulfilled for all the samples and if instrument reports are regular. Therefore, the evaluation report is produced, containing all information about participants, quality tests, results, target and performance evaluation.
Results
Participants
In the end a total of 62 previously authorized laboratories, 32 public and 30 private laboratories, equipped with different PCR systems validated, participated in three distribution rounds (EQA1/EQA2/EQA3) of the INSTAND Extra EQA scheme “Virus Genome Detection – SARS-CoV-2” (340). For SARS-CoV-2 molecular detection testing, the number of participating laboratories increased steadily: 41 registered laboratories in the first EQA round (EQA1), 54 in the second EQA round (EQA2) and 62 in the third EQA round (EQA3) (Figure 1).
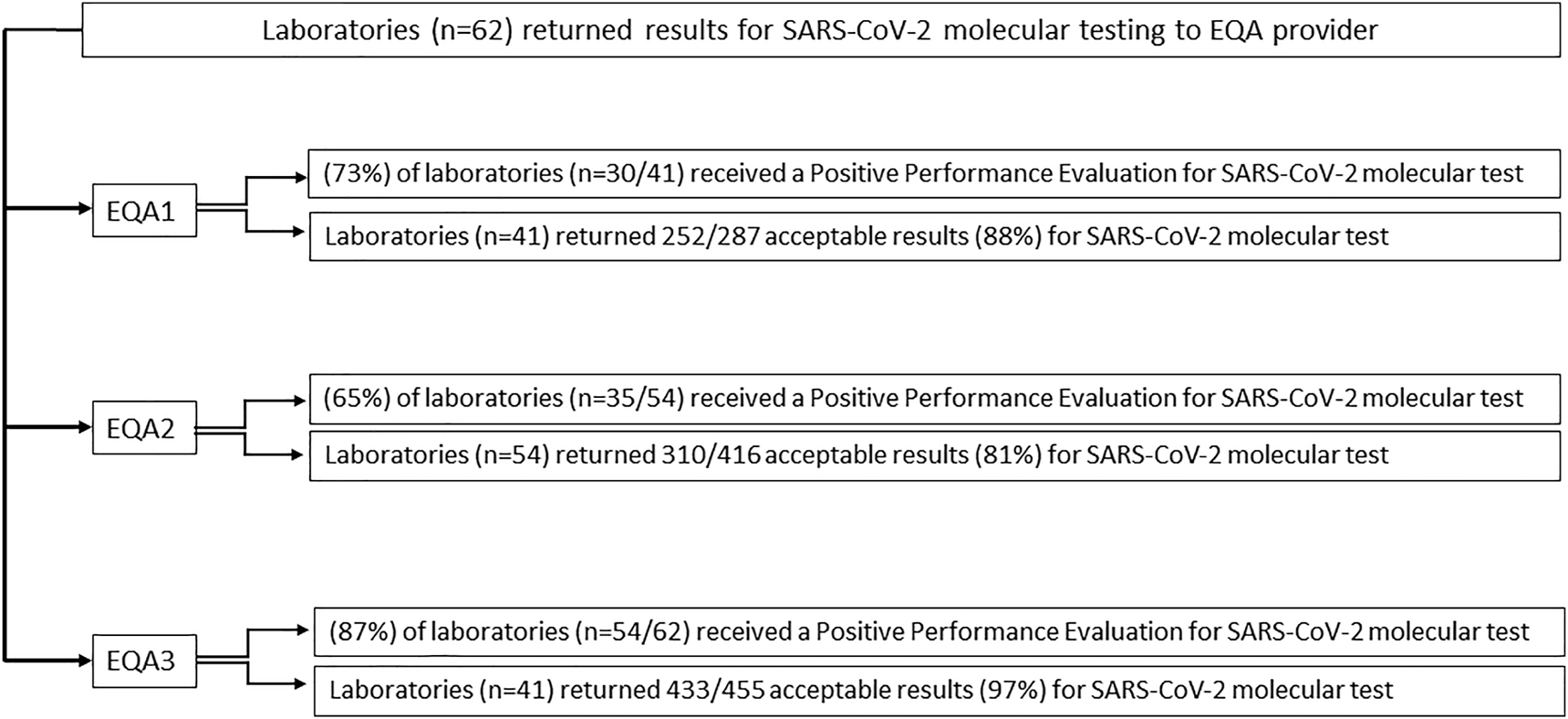
Positive performance evaluation: the flow diagram summarizes the performance evaluation results of sixty-two participating laboratories.
Overall SARS-CoV-2 molecular test performance
The blind work setting was necessary to avoid false laboratory results and so false reports by the participating authorized laboratories. The false certifications would have a devastating effect in the management of the pandemic, as the diagnosis for COVID-19, unlike what happens for many other diseases, is exclusively linked to the outcome of the laboratory investigation in molecular biology. The incorrect diagnosis for COVID-19 would also have important implications on vaccination cycles and the issuance of the green pass.
In the first EQA round conducted between April, 8th 2020 and April, 20th 2020, 30 laboratories out of 41 (73%) received a Positive Performance evaluation (Figure 2A) for the CRQ’s EQA scheme COVID437 (252 samples out of 287 processed (88%) received an acceptable evaluation).
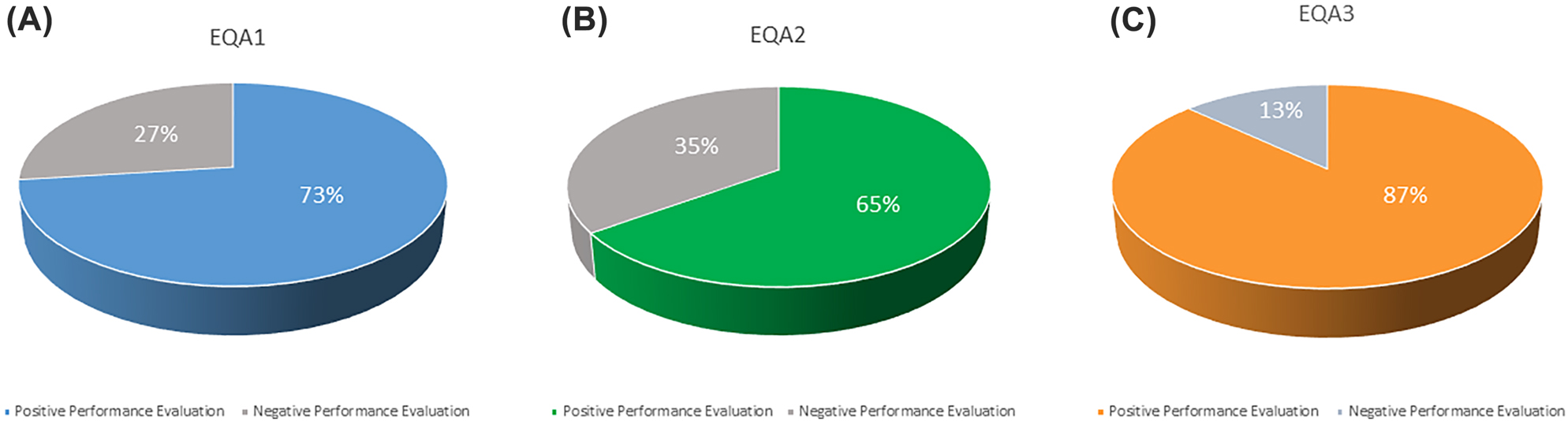
Overall performance of laboratories: the graphs show the average percentage of positive/negative evaluation of participating laboratories for each EQA rounds: (A) EQA1; (B) EQA2; (C) EQA3.
In the second EQA round conducted between July, 1st 2020 and July, 17th 2020, 35 laboratories out of 54 (65%) received a Positive Performance evaluation (Figure 2B) for the CRQ’s EQA scheme COVID437 (310 samples out of 416 processed (81%) received an acceptable evaluation). In the third EQA round conducted between November, 18th 2020 and November, 26th 2020, 54 laboratories out of 62 (87%) received a Positive Performance evaluation (Figure 2C) for the CRQ’s EQA scheme COVID437 (433 samples out of 455 processed (97%) received an acceptable evaluation). The laboratories that received a Negative Performance evaluation, (27% for EQA1, 35% for EQA2 and 13% for EQA3) (Figure 2A–C) have incorrectly interpreted the EQA exercise samples, either mistaking the interpretation of the instrumental data or the results compared to the expected outcomes (negative samples given as positive or positive samples given for negative).
The simulation test based on a real patient, carried out with the EQA program considered (7 samples, one of which was educational) and covered a total of 1,453 tests with 87.8, 74.5 and 95.2% agreement with the expected outcomes for the EQA1, EQA2 and EQA3 respectively (Figure 3A–C).
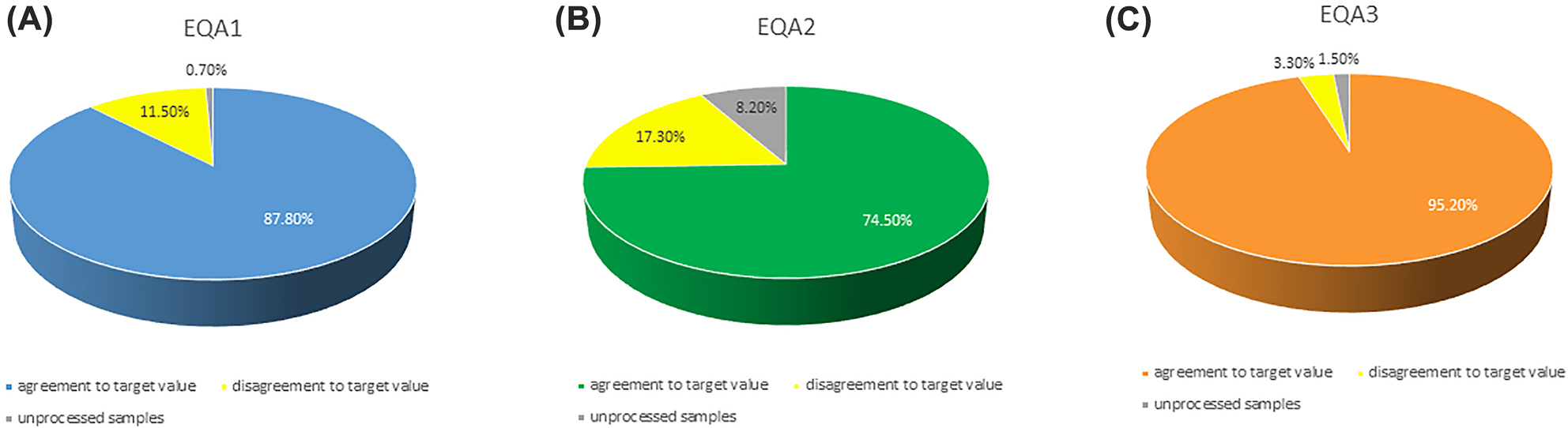
Performance on single sample: the graphs show the percentage of correct result (agreement to target value) of the final interpretations for each EQA rounds: (A) EQA1; (B) EQA2; (C) EQA3.
The selected test genes were: E, N, N1, N2, S, Orf1ab and RdRp. A successful participation means the achievement of 100% correct results in all the 7 samples according to the target values. In the EQA1 round the success rate for each sample test was respectively 92.7, 82.9, 82.9, 90.2, 85.4, 87.8, and 92.7% (Supplementary Table S1). In the EQA2 round the success rate for each sample test was respectively 61.1, 92.6, 92.6, 57.4, 87.0, 85.2 and 88.9% (Supplementary Table S2) and In the EQA3 round the success rate for each sample test was respectively 100, 97, 100, 89, 97, 95 and 89% (Supplementary Table S3). Interestingly, the 75% performed the SARS-CoV-2 detection with success, while the 25% failed either in one or all the 7 samples test.
The failure is mainly due to errors during the analysis (58.6%) probably due to contaminations, not-specific detection, or an operator error. Other reasons (41.4%) including inhibition of internal control, lower results due to the limit of detection (LOD), and incomplete results such as less of 7 EQA samples analyzed.
Nucleic acid extraction methods test performance
A total of 55 different nucleic acid extraction KIT (Table 1) were used. The most frequently used systems were QIAamp DSP Virus Spin Kit (QIAGEN Pty Ltd, Clayton, Australia), Magrev Viral DNA/RNA Extraction Kit (Anatolia GeneWorks, Sultanbeyli/İstanbul, Turkey) and STARMag 96x4 Universal Cartridge Kit (Seegene Inc, Walnut Creek, CA, USA), Xpert Xpress SARS-CoV- 2 (Cepheid, Sunnyvale, CA, USA).
List of the most utilized extraction methods and platforms.
| Extraction methods used | All samples correct | Total samples | % Correct |
|---|---|---|---|
| Anatolia GeneWorks | 112 | 127 | 86% |
| BioMerieux – easyMAG | 39 | 39 | 100% |
| Cepheid – GeneXpert | 96 | 102 | 94% |
| DiaSorin S.p.A. – Simplexa | 83 | 83 | 100% |
| Norgen Biotek | 85 | 90 | 93% |
| Promega Corporation – Maxwell | 71 | 74 | 98% |
| QIAGEN – QIAamp | 281 | 300 | 96% |
| RBC Bioscience – MagCore | 94 | 103 | 91% |
| Roche Diagnostics – MagNA | 93 | 97 | 97% |
| Seegene Inc. – STARMag | 104 | 106 | 93% |
| Shanghai ZJ Bio-Tech Co | 103 | 111 | 89% |
| Thermo Fisher Scientific – MagMAX | 91 | 97 | 96% |
| Enbiotech – SARS-COV-2 | 25 | 32 | 78% |
| Fast Track Diagnostics – SARS-CoV-2 | 22 | 26 | 85% |
| Hain Lifescience GmbH – GTX Extraction kits | 24 | 25 | 96% |
| Hangzhou Bioer Technology – MagaBio plus Virus | 33 | 35 | 94% |
| Liferiver – Viral RNA Isolation Kit | 15 | 15 | 100% |
| Menarini Diagnostics – Test VitaPCRTM SARS-CoV-2 | 23 | 57 | 40% |
| Molgen – PurePrep Pathogens | 13 | 14 | 93% |
| Siemens Healthcare – VERSANT Sample | 22 | 26 | 85% |
| TIB MOLBIOL GmbH – LightMix Modular Wuhan CoV RdRP-gene | 11 | 12 | 92% |
| Zymo Research – Quick-RNA Viral Kit | 22 | 24 | 92% |
| Other | 49 | 56 | 87% |
Detection assays
A total of 55 different amplification SARS-CoV-2 kit platforms (Table 2) were used The most frequently used system were Allplex 2019 n-CoV Assay (Seegene Inc. Walnut Creek, CA, USA), TaqPath COVID-19 (Thermo Fisher Scientific, Waltham, MA, USA), Novel Coronavirus (2019-nCoV) Real Time RT-PCR Kit (Shanghai ZJ Bio-Tech Co, Shanghai, China), and COVID-19 HT Screen (CLONIT srl, Siziano PV, Italy). It is important to underline that, among the different PCR systems used by the participating laboratories, there was one PCR assay, VitaPCRTM SARS-CoV-2 from Menarini Diagnostics (Ripoli, FI, Italy), with the absence of an RNA extraction separated step, has demonstrated to be less sensitive than the other tests. Indeed, the laboratories that used this particular test have completely failed the EQA1 and EQA2 rounds, giving an indeterminate result. Only two participants among 62 used this direct assay, but considering that the direct test was used also in emergency areas and islands, in order to avoid report inhomogeneity, subsequently, a specific EQA scheme was implemented for this type of assay.
List of the most utilized PCR reagents and kit.
| - | All samples correct | Total samples | % Correct |
|---|---|---|---|
| Adaltis – MOLgen SARS-CoV-2 | 78 | 82 | 97% |
| Anatolia GeneWorks – Novel Coronavirus | 62 | 77 | 78% |
| Cepheid – Xpert Xpress SARS-CoV-2 | 97 | 103 | 94% |
| CLONIT- HT Screen | 110 | 121 | 89% |
| GeneMatrix – NeoPlex COVID-19 | 49 | 54 | 91% |
| Imegen – SARS-CoV-2 | 11 | 12 | 92% |
| Hangzhou Bioer Technology | 29 | 29 | 100% |
| Norgen Biotek – 2019-nCov TaqMan | 64 | 68 | 97% |
| Nuclear Laser Medicine – SARS CoV-2 Real Time | 26 | 26 | 100% |
| OSANG Healthcare | 27 | 30 | 90% |
| Roche Diagnostics – GENEMATRIX | 27 | 28 | 96% |
| Roche Diagnostics – LightMix Modular SARS | 74 | 78 | 96% |
| Sacace Biotechnologies – SARS-CoV-2 | 11 | 14 | 79% |
| Seegene – Allplex 2019 n-CoV Assay | 285 | 292 | 99% |
| Shanghai Kehua Bio-engeneering | 29 | 36 | 81% |
| Shanghai ZJ Bio-Tech Co | 108 | 115 | 94% |
| Thermo Fisher Scientific – TaqPath | 121 | 122 | 99% |
| TIB MOLBIOL GmbH – LightMix Modular SARS | 22 | 24 | 92% |
| Vitassay Healthcare – Vitassay | 19 | 19 | 100% |
| Wuhan HealthCare Biotechnology | 19 | 19 | 100% |
| Other | 58 | 64 | 92% |
Discussion
Hereby, we report the first Italian EQA scheme for SARS-CoV-2 molecular detection assigned to clinical laboratories in Sicily during the 2020. In particular, 62 Sicilian laboratories (32 public and 30 private) were involved. It is important to note that these laboratories had previously been authorized, from the Regional Government to perform COVID-19 case identification, based on specific, structural, technologic and professional features. The laboratories participated in the EQA following the three distribution rounds (EQA1/EQA2/EQA3) scheme.
Seven EQA samples were shipped to each participant laboratory. In order to fulfil the quality criteria and to overcome to technical issues, the 7 test samples (E, N, N1, N2, S, Orf1ab and RdRp) were subministrated to the laboratories in a blind and name-inverted way.
Interestingly, the overall results of SARS-CoV-2 molecular test 2020 performance showed that 75% of the laboratories received a positive performance evaluation while 25% did not receive a good score but a negative performance evaluation. The percentage of laboratories that received a positive performance evaluation for each EQA1/EQA2/EQA3 were respectively 73%/65%/87%.
Within the population (25% of the participants) who received a negative performance evaluation, it is observed that the test failure is mainly due to errors during the analysis (58.6%), probably on account of contaminations, not-specific detection, or an error caused by the operator. Other reasons (41.4%), including inhibition of internal control, lower results due to the limit of detection (LOD), and incomplete results, such as less than 7 EQA samples analyzed.
Among the public and private laboratories, the most used nucleic acid extraction methods and platforms were: QIAamp DSP Virus Spin Kit (QIAGEN Pty Ltd, Clayton, Australia), Magrev Viral DNA/RNA Extraction Kit (Anatolia GeneWorks, Sultanbeyli/İstanbul, Turkey) and STARMag 96x4 Universal Cartridge Kit (Seegene Inc, Walnut Creek, CA, USA), and Xpert Xpress SARS-CoV- 2 (Cepheid). Whereas, the most used detection assays were: Allplex 2019 n-CoV Assay (Seegene Inc. Walnut Creek, CA, USA), TaqPath COVID-19 (Thermo Fisher Scientific, Waltham, MA, USA), Novel Coronavirus (2019-nCoV) Real Time RT-PCR Kit (Shanghai ZJ Bio-Tech Co, Shanghai, China), and COVID-19 HT Screen (CLONIT srl, Siziano PV, Italy).
The early and successful detection of SARS-CoV-2 in likely patients is the crucial element for reducing the viral transmission among the population. The government strategies focused on diagnosis, isolation and contact tracing. For this reason, the diagnostic tests able to drive the aforementioned approaches are a matter of global health deal. In order to give a significant contribution to the management of the SARS-CoV-2 pandemic, the reliability of the molecular results and so, the outcomes of the EQA, were fundamental elements, not only for the usual processes of improvement of overall performance of the individual laboratories, but mainly for the maintenance of the authorization to carry out laboratory activities for the diagnosis and certification of the COVID-19 disease.
The blind work setting is always necessary to avoid false laboratory results and therefore false reports by the participating and authorized laboratories. The false certifications have a devastating effect on any healthcare processes, especially in the management of a pandemic such as COVID-19, unlike what happens with other diseases. The incorrect diagnosis for COVID-19, exclusively linked to the outcome of the laboratory investigation in molecular biology, has important implications on the vaccination cycles and the issuance of the Green Pass.
For these reasons, “the authorization to carry out molecular biology investigation for the research of the SARS-CoV-2 virus was suspended in the population (25% of the participants) that received a negative performance evaluation in any sample of the EQA scheme, as well was suspended the authorization to issue reports diagnosis of COVID-19 disease”.
The participation and the positive performance of specific and related EQA schemes are required, by the current legislation, for the accreditation of laboratories, together with various guidelines and international technical standards of the sector (ISO/IEC 17025, ISO 15189, Joint Commission, etc.) [14, 15].
The actual study performed in Italy, has implemented an EQA scheme for COVID-19, and was organized in a blind, centralized and governmental way. Our study allowed us to identify pockets of non-compliance that the normal health governance processes had not been able to highlight. The results of the study suggest that, the EQA schemes should become a valid standard of authorization, accreditation and a goal strategy for continuous verification of the possession of the requirements so as to operate in the name of the service of patients and on behalf of the Health Services.
Acknowledgments
We thank Ing. Mario la Rocca, General Manager of the “Dipartimento Pianificazione Strategica dell’Assessorato della Salute di Palermo”.
-
Research funding: None declared.
-
Author contributions: FDG, GB and FC conceived the study. FDG, GB and AG wrote the manuscript. GB and FC prepared, performed and analysed the samples and results of the EQA rounds. FC run the statistical analysis. All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Competing interests: Authors state no conflict of interest.
-
Informed consent: Not applicable.
-
Ethical approval: Not applicable.
References
1. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---15-june-2022.Search in Google Scholar
2. Available from: https://covid19.who.int/region/euro/country/it.Search in Google Scholar
3. Budinger, GRS, Misharin, AV, Ridge, KM, Singer, BD, Wunderink, RG. Distinctive features of severe SARS-CoV-2 pneumonia. J Clin Invest 2021;131:e149412. https://doi.org/10.1172/jci149412.Search in Google Scholar
4. Wu, F, Zhao, S, Yu, B, Chen, YM, Wang, W, Song, ZG, et al.. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265–9. https://doi.org/10.1038/s41586-020-2008-3.Search in Google Scholar PubMed PubMed Central
5. Montgomery, L, Macy, JM. Characterization of rat cecum cellulolytic bacteria. Appl Environ Microbiol 1982;44:1435–43. https://doi.org/10.1128/aem.44.6.1435-1443.1982.Search in Google Scholar PubMed PubMed Central
6. Chan, JF, Kok, KH, Zhu, Z, Chu, H, To, KK, Yuan, S, et al.. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microb Infect 2020;9:221–36. https://doi.org/10.1080/22221751.2020.1719902.Search in Google Scholar PubMed PubMed Central
7. World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance. Geneva: World Health Organization; 2020.Search in Google Scholar
8. Lippi, G, Favresse, J, Gromiha, MM, SoRelle, JA, Plebani, M, Henry, BM. Ad interim recommendations for diagnosing SARS-CoV-2 infection by the IFCC SARS-CoV-2 variants working group. Clin Chem Lab Med 2022;60:975–81. https://doi.org/10.1515/cclm-2022-0345.Search in Google Scholar PubMed
9. Service, RF. A call for diagnostic tests to report viral load. Science 2020;370:22. https://doi.org/10.1126/science.370.6512.22.Search in Google Scholar PubMed
10. Buchta, C, Görzer, I, Chiba, P, Camp, JV, Holzmann, H, Puchhammer-Stöckl, E, et al.. Variability of cycle threshold values in an external quality assessment scheme for detection of the SARS-CoV-2 virus genome by RT-PCR. Clin Chem Lab Med 2020;59:987–94. https://doi.org/10.1515/cclm-2020-1602.Search in Google Scholar PubMed
11. Cuypers, L, Bode, J, Beuselinck, K, Laenen, L, Dewaele, K, Janssen, R, et al.. Nationwide harmonization effort for semi-quantitative reporting of SARS-CoV-2 PCR test results in Belgium. Viruses 2022;14:1294. https://doi.org/10.3390/v14061294.Search in Google Scholar PubMed PubMed Central
12. Lippi, G, Plebani, M. The many clinical advantages of reporting the cycle threshold (Ct) value. Ann Transl Med 2022;10:427. https://doi.org/10.21037/atm-22-1104.Search in Google Scholar PubMed PubMed Central
13. ISO/IEC 17043. Conformity assessment—General requirements for proficiency testing providers. Geneva: ISO; 2010.Search in Google Scholar
14. ISO/IEC 17025. General requirements for the competence of testing and calibration laboratories. Geneva: ISO; 2017.Search in Google Scholar
15. ISO 15189. Medical laboratories—Requirements for quality and competence. Geneva: ISO; 2012.Search in Google Scholar
Supplementary Material
The online version of this article offers supplementary material (https://doi.org/10.1515/cclm-2022-0780).
© 2022 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorial
- A new milestone on the road to global standardization of apolipoprotein measurements
- Review
- Saliva – a new opportunity for fluid biopsy
- Opinion Papers
- Emerging technology: a definition for laboratory medicine
- The next wave of innovation in laboratory automation: systems for auto-verification, quality control and specimen quality assurance
- EFLM Paper
- The new, race-free, Chronic Kidney Disease Epidemiology Consortium (CKD-EPI) equation to estimate glomerular filtration rate: is it applicable in Europe? A position statement by the European Federation of Clinical Chemistry and Laboratory Medicine (EFLM)
- Guidelines and Recommendations
- Overcoming challenges regarding reference materials and regulations that influence global standardization of medical laboratory testing results
- General Clinical Chemistry and Laboratory Medicine
- Quantitative protein mass-spectrometry requires a standardized pre-analytical phase
- Report from the HarmoSter study: inter-laboratory comparison of LC-MS/MS measurements of corticosterone, 11-deoxycortisol and cortisone
- The influence of proteoforms: assessing the accuracy of total vitamin D-binding protein quantification by proteolysis and LC-MS/MS
- Total serum vitamin B12 (cobalamin) LC-MS/MS assay as an arbiter of clinically discordant immunoassay results
- The preanalytical process in the emergency department, a European survey
- Proenkephalin A as a marker for glomerular filtration rate in critically ill children: validation against gold standard iohexol GFR measurements
- Umbilical cord blood gases: probability of arterial or venous source in acidemia
- Reference Values and Biological Variations
- Pediatric reference interval verification for 17 specialized immunoassays and cancer markers on the Abbott Alinity i system in the CALIPER cohort of healthy children and adolescents
- Hematology and Coagulation
- Performance of digital morphology analyzer CellaVision DC-1
- Cancer Diagnostics
- Managing the impact of inter-method bias of prostate specific antigen assays on biopsy referral: the key to move towards precision health in prostate cancer management
- Cardiovascular Diseases
- Quantitation of cardiac troponin I in cancer patients treated with immune checkpoint inhibitors: a case-control study
- Infectious Diseases
- A novel scoring system combining Modified Early Warning Score with biomarkers of monocyte distribution width, white blood cell counts, and neutrophil-to-lymphocyte ratio to improve early sepsis prediction in older adults
- Technical and health governance aspects of the External Quality Assessment Scheme for the SARS-CoV-2 molecular tests: institutional experience performed in all clinical laboratories of a Regional Health Service
- Acknowledgment
- Acknowledgment
- Letters to the Editor
- About the estimation of albuminuria based on proteinuria results
- Response to “About the estimation of albuminuria based on proteinuria results”
- Reply to Abildgaard et al.: lot variation and inter-device differences contribute to poor analytical performance of the DCA vantage™ HbA1c POCT instrument in a true clinical setting
- Reply to letter from Mayfield et al. regarding “Lot variation and inter-device differences contribute to poor analytical performance of the DCA Vantage™ HbA1c POCT instrument in a true clinical setting”
- High-sensitive cardiac troponin T: are turbulences coming?
- Analytical performance evaluation of bioactive adrenomedullin on point-of-care platform
- Increased PD-L1 surface expression on peripheral blood granulocytes and monocytes after vaccination with SARS-CoV2 mRNA or vector vaccine
- Neopterin level can be measured by intraocular liquid biopsy
- The stability of pleural fluid pH under slushed ice and room temperature conditions
Articles in the same Issue
- Frontmatter
- Editorial
- A new milestone on the road to global standardization of apolipoprotein measurements
- Review
- Saliva – a new opportunity for fluid biopsy
- Opinion Papers
- Emerging technology: a definition for laboratory medicine
- The next wave of innovation in laboratory automation: systems for auto-verification, quality control and specimen quality assurance
- EFLM Paper
- The new, race-free, Chronic Kidney Disease Epidemiology Consortium (CKD-EPI) equation to estimate glomerular filtration rate: is it applicable in Europe? A position statement by the European Federation of Clinical Chemistry and Laboratory Medicine (EFLM)
- Guidelines and Recommendations
- Overcoming challenges regarding reference materials and regulations that influence global standardization of medical laboratory testing results
- General Clinical Chemistry and Laboratory Medicine
- Quantitative protein mass-spectrometry requires a standardized pre-analytical phase
- Report from the HarmoSter study: inter-laboratory comparison of LC-MS/MS measurements of corticosterone, 11-deoxycortisol and cortisone
- The influence of proteoforms: assessing the accuracy of total vitamin D-binding protein quantification by proteolysis and LC-MS/MS
- Total serum vitamin B12 (cobalamin) LC-MS/MS assay as an arbiter of clinically discordant immunoassay results
- The preanalytical process in the emergency department, a European survey
- Proenkephalin A as a marker for glomerular filtration rate in critically ill children: validation against gold standard iohexol GFR measurements
- Umbilical cord blood gases: probability of arterial or venous source in acidemia
- Reference Values and Biological Variations
- Pediatric reference interval verification for 17 specialized immunoassays and cancer markers on the Abbott Alinity i system in the CALIPER cohort of healthy children and adolescents
- Hematology and Coagulation
- Performance of digital morphology analyzer CellaVision DC-1
- Cancer Diagnostics
- Managing the impact of inter-method bias of prostate specific antigen assays on biopsy referral: the key to move towards precision health in prostate cancer management
- Cardiovascular Diseases
- Quantitation of cardiac troponin I in cancer patients treated with immune checkpoint inhibitors: a case-control study
- Infectious Diseases
- A novel scoring system combining Modified Early Warning Score with biomarkers of monocyte distribution width, white blood cell counts, and neutrophil-to-lymphocyte ratio to improve early sepsis prediction in older adults
- Technical and health governance aspects of the External Quality Assessment Scheme for the SARS-CoV-2 molecular tests: institutional experience performed in all clinical laboratories of a Regional Health Service
- Acknowledgment
- Acknowledgment
- Letters to the Editor
- About the estimation of albuminuria based on proteinuria results
- Response to “About the estimation of albuminuria based on proteinuria results”
- Reply to Abildgaard et al.: lot variation and inter-device differences contribute to poor analytical performance of the DCA vantage™ HbA1c POCT instrument in a true clinical setting
- Reply to letter from Mayfield et al. regarding “Lot variation and inter-device differences contribute to poor analytical performance of the DCA Vantage™ HbA1c POCT instrument in a true clinical setting”
- High-sensitive cardiac troponin T: are turbulences coming?
- Analytical performance evaluation of bioactive adrenomedullin on point-of-care platform
- Increased PD-L1 surface expression on peripheral blood granulocytes and monocytes after vaccination with SARS-CoV2 mRNA or vector vaccine
- Neopterin level can be measured by intraocular liquid biopsy
- The stability of pleural fluid pH under slushed ice and room temperature conditions

