Abstract
Pluripotent stem cells (PSCs), characterized by self-renewal and capacity of differentiating into three germ layers, are the programmable building blocks of life. PSC-derived cells and multicellular systems, particularly organoids, exhibit great potential for regenerative medicine. However, this field is still in its infancy, partly due to limited strategies to robustly and precisely control stem cell behaviors, which are tightly regulated by inner gene regulatory networks in response to stimuli from the extracellular environment. Synthetic receptors and genetic circuits are powerful tools to customize the cellular sense-and-response process, suggesting their underlying roles in precise control of cell fate decision and function reconstruction. Herein, we review the progress and challenges needed to be overcome in the fields of PSC-based cell therapy and multicellular system generation, respectively. Furthermore, we summarize several well-established synthetic biology tools and their applications in PSC engineering. Finally, we highlight the challenges and perspectives of harnessing synthetic biology to PSC engineering for regenerative medicine.
Introduction
Stem cells are cells capable of self-renewing and differentiating into multiple lineages. During embryonic development, stem cells can differentiate into presumptive lineage cells and further organize into complex organs. According to the different developmental potency, stem cells can generally be divided into four categories [1], which are totipotent stem cells, pluripotent stem cells (PSCs), multipotent stem cells, and unipotent stem cells (Figure 1). Totipotent stem cells containing fertilized eggs and blastomeres at early cleavage stage have been thought to possess the highest differentiation potential to form both embryonic and extra-embryonic tissues. After several round of divisions, the totipotent stem cells form a blastocyst, containing trophoblast and an inner cell mass (ICM) which will further differentiate into the epiblast and the primitive endoderm, respectively. The epiblast cells are PSCs that can differentiate into any cell type of the embryo during development. Compared with totipotent stem cells, PSCs exhibit less differentiation capacity since they can develop into the three germ layers of the embryos but not the cells of extra-embryonic tissues (Figure 1). Two milestones of human stem cell technologies were the successful establishment of embryonic stem cell (ESC) line from human blastocysts in 1998 [2], and the generation of induced pluripotent stem cells (iPSCs) from human adult fibroblast via transduction of certain transcription factors (TFs) in 2007 [3, 4]. Both ESCs and iPSCs belong to PSCs for their unlimited capability of self-renewal and differentiation into the three germ layers. After a series of lineage commitment events, PSCs give rise to multipotent stem cells with the potential to differentiate into certain lineages, such as hematopoietic stem cells (HSCs) (Figure 1). In addition, unipotent stem cells display very limited developmental potency and can only differentiate into one specific type of cell (Figure 1).
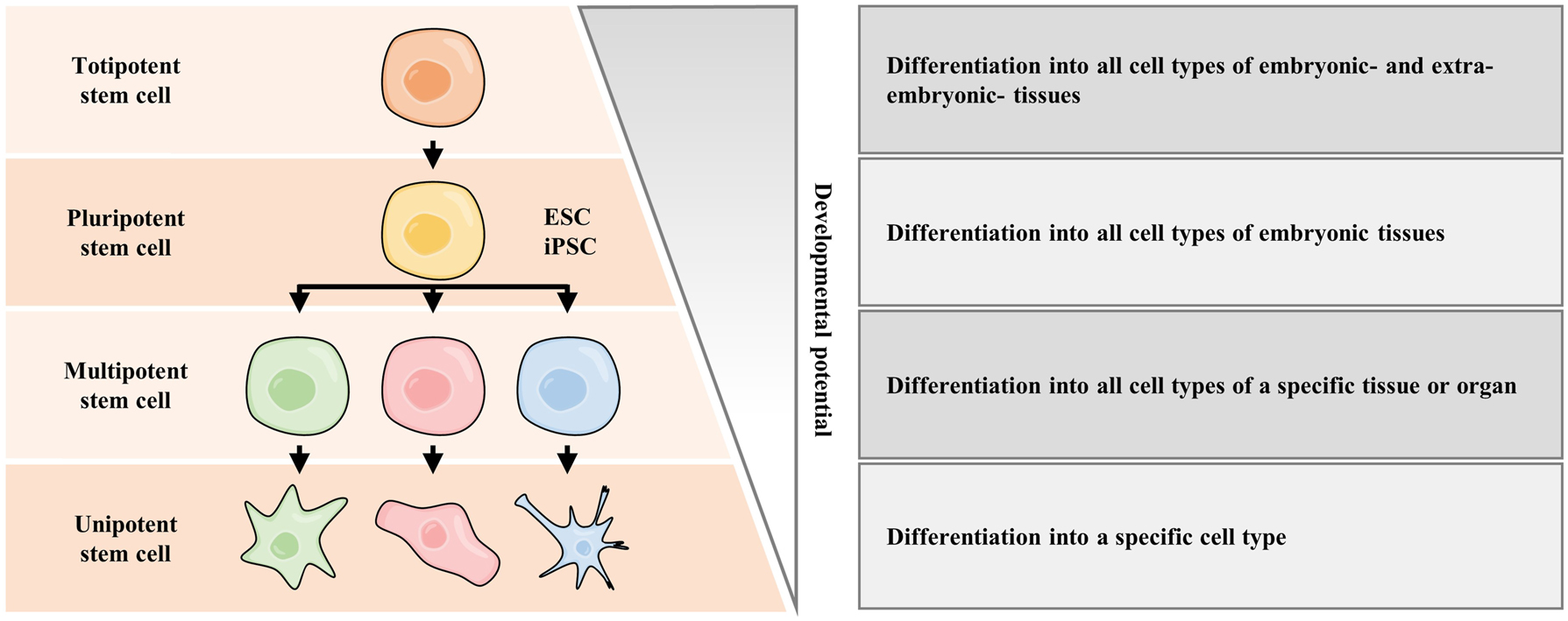
Stem cells with diverse developmental potential. Stem cells are classified by their potency to differentiate into specialized cell types. The most potent stem cell is the totipotent stem cell, including the fertilized eggs. Totipotent stem cell can differentiate into inner cell mass, which is ESCs. Notably, both ESCs and iPSCs belong to pluripotent stem cells. As development progresses, pluripotent stem cells experience lineage commitment and become multipotent stem cells. The least potent one is the unipotent stem cell. ESC, embryonic stem cell; iPSC, induced pluripotent stem cell.
Due to the outstanding properties mentioned above, human PSCs (hPSCs) have exhibited prominent applications in regenerative medicine, of which the ultimate goal is to reverse dysfunctional tissues to healthy conditions with full functions by repairing, replacing, or even in situ regenerating the cells/organs of patients.
The application of stem cells and their derivatives in cell therapy has been investigated for several decades but is still at an early stage. The most acceptable and curative cell therapy is HSC transplantation (HSCT) for acute myeloid leukemia, proposed by Edward Donnall Thomas, who won the Nobel Prize in Physiology or Medicine in 1990 for his outstanding work on bone marrow transplantation [5]. Apart from HSCs, several other types of stem cells have been used for pre-clinical studies and clinical treatment, such as PSC-derived cells. To date, according to ClinicalTrials.gov (key words are “stem cells”), there are 7,780 cases of stem cell-based clinical trials registered worldwide, targeting to treat various types of diseases containing musculoskeletal, neurology and pneumology of COVID-19. Meanwhile, PSC-based cell therapies are mostly under pre-clinical study. So far, there are approximately 130 cases of PSC-based clinical trials (ClinicalTrials.gov, key words are “embryonic stem cells” and “induced pluripotent stem cells”), including Parkinson’s Disease (PD), macular degeneration, retinitis pigmentosa, corneal disorder, heart failure, and spinal cord injury (SCI).
However, several limitations hamper the clinical translation of PSC-based cell therapy, including immune rejection; tumorigenicity; robust and homogeneous differentiation of specific cell types that can survive, proliferate, and function maturely; inefficiency of scale-up and quality control for cell manufacture, which need to be addressed urgently.
In addition to hPSC-based cell therapy, hPSCs have been used to generate multicellular tissues in vitro, aiming to serve as a tissue or organ-level source for organ repair and regeneration in the future. The fast developing stem cell technologies, such as organoid technology and synthetic embryo technology, allow stem cells to generate sophisticated multicellular systems via simulating embryological processes in vitro. By using various cocktails of small-molecule compounds to mimic in vivo developmental trajectories, hPSC-derived organoids have spanned various types of organs derived from ectoderm (brain [6] and retina [7] organoids), endoderm (lung [8], gastric [9], colon [10], intestine [11], liver [12] and pancreatic [13] organoids), and mesoderm (heart [14], kidney [15] and blood [16] organoids), respectively. Besides, a wave of articles has recently reported the successful generation of synthetic human blastoids, modeling pre-implantation blastocyst development and even implantation [17], [18], [19], [20], [21], [22], [23]. However, more efforts must be made to improve these multicellular systems to fulfill the transplantation criteria, including satisfactory functions, suitable tissue sizes, better immune tolerance, low cost, and high reproducibility.
To accelerate the development of PSC-based regenerative medicine, the cooperation of multiple disciplines is necessary to address its challenges, such as 3D-bioprinting technology, system biology, and synthetic biology. Notably, different from traditional top-down strategies to investigate developmental events and regulate cellular functions, the theoretical rationale of synthetic biology, which is bottom-up construction of systems, provides a novel perspective to realize sophisticated, programmable control of cellular behaviors. Synthetic biology was initially explored in bacteria and yeast due to their relatively simple and clear genetic regulatory networks. In 2000, the first bistable switch in Escherichia coli was established, achieving controllable switching between two stable gene expression states in engineered cells [24]. In the same year, scientists designed an oscillator, a gene circuit composed of a triple negative feedback loop, which achieved a periodic oscillation of repressor protein expression [25]. These two findings have marked the beginning of an “era of synthetic biology”. In 2002, another group used a set of transcriptional regulators and their corresponding promoters to construct genetic circuits that exhibited the properties of binary logic gates, laying the groundwork for the implementation of complex Boolean logic computations within cells [26]. By further refining and combining these basic elements or by reforming natural sense-response systems, more complex genetic circuits that could execute certain functions, such as bandpass filters [27] and counters [28] were developed. With the fast development of gene editing techniques and increasing understanding of cell biology, the engineered objects of synthetic biology have shifted from predominantly bacteria and yeast towards mammalian cells [29], indicating its promising application in stem cell technologies to precisely regulate the states and functions of PSCs and their derivatives.
Herein, this review will mainly emphasize the important impacts and underlying applications of synthetic biology in PSC bioengineering for facilitating and fulfilling the needs of regenerative medicine. We will review the applications of PSCs in regenerative medicine and discuss the limitations that need to be addressed. Then we will introduce the important synthetic biology tools in mammalian cells and illustrate how they facilitate the development of PSC-based regenerative medicine. Finally, we will discuss the challenges and perspectives of engineering PSCs for regenerative medicine by using synthetic biology.
Applications of PSCs in regenerative medicine
Due to the outstanding characteristics of hPSCs mentioned in the Introduction, they have been regarded as the better “chassis” for the applications of regenerative medicine by establishing pools of various types of cells for cell therapy or generating multicellular systems for organ transplantation in the future.
Progress of PSC-derived cell therapies
hPSCs and their derivatives have been applied in cell therapy for several intractable diseases, and a considerable number of products based on hPSCs have entered pre-clinical and clinical research. One of the first clinical trials of hPSC-based cell therapy was conducted in 2010, which used human ESC (hESC)-derived oligodendrocyte progenitor cells to treat SCI (NCT01217008). Subsequently, in 2012, two prospective clinical studies using the hESC-derived retinal pigment epithelium (RPE) cells to treat Stargardt’s macular dystrophy and dry age-related macular degeneration were launched (NCT01345006 and NCT01344993). The two trials have demonstrated the medium-term to long-term safety and graft survival of the transplanted cells, as well as their possible biological activity [30]. Besides, in 2014, researchers started the first clinical trial using iPSC-derived cells to treat eye diseases [31]. They injected iPSC-derived RPE cells into a patient suffering from age-related macular degeneration. Unfortunately, the trial was halted in 2015 for the concern of the potential tumorigenesis of two small genetic mutations in the cells. In addition, hPSCs have also been used for treating heart diseases. The safety of hPSC-derived cardiac-committed progenitor cells was evaluated after being seeded in fibrin gel scaffolds and injected into 10 patients with severe heart failure (phase I trial; ESCORT, NCT02057900) [32]. The trial demonstrated the feasibility of generating clinical-grade hESC-derived cardiomyocyte progenitors. One patient died from unrelated comorbidities. At the 1-year endpoint, all the other patients had uneventful recoveries, including a significant enhancement of the systolic motion of the treated segments. With a maximum follow-up of 6 years, no patient presented an adverse event related to the cells and/or the patch. Another patient died from heart failure 22 months after treatment [32]. Recently, another clinical trial was carried out to assess the safety of human iPSC (hiPSC)-derived cardiomyocytes embedded in a cell sheet for transplantation (jRCT2052190081). Among PSC-based cell therapies for neural diseases, one of the most popular is hPSC-derived dopaminergic neurons replacement for PD. In 2017, scientists led the first ESC-based phase I/IIa clinical study of PD in China (NCT03119636) [33]. This study assessed the safety and efficacy of intracerebral transplantation of hESC-derived neural precursor cells in patients with PD.
Until October 15, 2023, approximately 130 clinical studies based on hPSC derivatives have been registered, according to ClinicalTrials.gov (key words: embryonic stem cells and induced pluripotent stem cells). The clinical researches of hiPSC-derived cells cover PD, macular degeneration, retinitis pigmentosa, corneal disorder, heart failure, SCI, platelet transfusion, graft-versus-host diseases, cartilage defects, and cancers; while the clinical researches of hESC-derived cells involve PD, macular degeneration, retinitis pigmentosa, amyotrophic lateral sclerosis, SCI, Type I diabetes, citrullinemia type I, Intrauterine adhesions.
Despite the fact that PSCs have been used on a certain scale in clinical trials, there are still many challenges that need to be addressed for better application of PSCs to cell therapy, including: (i) immune rejection; (ii) tumorigenicity; (iii) robust and homogeneous differentiation of specific cell types that can survive, proliferate, and function maturely; (iv) inefficiency of scale-up and quality control for cell manufacture and so forth. Details about these problems and efforts to address them are as follows.
Immune rejection
One of the major safety concerns that need to be addressed prior to widespread clinical use of PSCs and their derivatives is immunogenicity [34]. Immune rejection is a major obstacle for allogeneic cell transplantation, mainly due to human leukocyte antigen (HLA) mismatch between the transplanted cells and the recipient. Besides HLA mismatch, other reasons include immune antigens presentation by PSCs due to prolonged in vitro culture, atypical antigens formation due to genomic mutations, incomplete reprogramming of somatic cells during iPSC reprogramming, and immune immaturity of PSC-derived cells [34]. Autologous, patient-specific iPSC-derived cells may overcome these challenges. However, autologous iPSCs are too costly given the necessary quality control and too slow to meet acute needs. An alternative strategy is to use HLA-matched, quality-controlled iPSC banks that are being developed in Japan [34]. Such banks could provide standardized sources of cells for clinical applications, overcoming the need for individualized autologous cell lines and addressing cost and scalability concerns. An additional approach is HLA cloaking, which involves inactivating HLA genes and/or overexpressing immunosuppressive factors in allogeneic stem cells [35, 36]. This approach can reduce the immunogenicity of allogeneic stem cells, making them more suitable as “off-the-shelf” cell therapy products for large-scale clinical applications. Only a very small number of hPSC lines or even one single line could be required to serve as a universal and standardized source of cells covering the entire world population. Such universal hPSCs would be allogeneic, more homogeneous, cheaper, and safer, making them better candidates for engineering and serving as off-the-shelf cell therapy products for large-scale clinical applications.
Tumorigenicity
Self-renewal ability is one of the important properties of hPSCs, but also a double-edged sword since its potential tumorigenesis if the cells continue to proliferate indefinitely after transplantation [34]. Besides, progenitor cells used for cell therapy also have been reported to exhibit the tumorigenic potential after transplantation. In addition, tumorigenesis can be caused by several other reasons, including cells with unexpected high proliferation capacity, genomic abnormalities due to long-term culture and gene-editing manipulations. Recently, a systematic study pointed out that over 20 % of both hPSCs and their in vitro derivatives harbored cancer-related mutation (s). Particularly, 64 % of these samples contained mutations in the TP53 gene, which is a common tumor suppressor gene and its dysregulation is highly correlated to tumorigenesis and cancer progression [37]. These results highlight the critical importance of vigilantly monitoring for cancer-related mutations in hPSCs, especially when they are being considered for clinical use. Since the potential tumorigenicity of hPSCs and their derivatives still cannot be completely ablated in vitro by genetic engineering, dynamically monitoring and eliminating the cells with tumorigenic transition is a practical approach to ensure the safety of the cell therapy product. For example, stem cells can be engineered with inducible suicide or elimination switches, such as fail-safe systems based on suicide genes Caspase9 and HSVtk [38, 39] and miRNA switches [40].
Robust and homogeneous differentiation of specific cell types that can survival, proliferate and function maturely
The differentiation of PSCs into desired cell types is a primary step in the development of cell-based therapies. However, there are significant challenges associated with the low efficiency and reproducibility of this process, resulting in generating cells with low purity, immature function and limited reproducibility. Additionally, for cell replacement therapy, transplanted cells must possess the sufficient survival and proliferation capacity, followed by homing and localizing to the therapeutic site and executing the therapeutic functions. Importantly, donor cells are hard to communicate and integrate with recipient tissues to initiate pre-designed functions. Taken the clinical trials of PSC-derived RPE cells to treat age-related macular degeneration (AMD) as an example, the transplanted RPE cells failed to precisely home and localize to the therapeutic site. Another example is the PSCs-derived dopaminergic progenitor cells-based PD therapy, the major challenges are durability and survival of cells, synaptic integration and functional restoration after transplantation [41].
Inefficiency of scale-up and quality control for cell manufacture
Similar to the manufacture of other biologics, that of hPSCs and their derivatives should be strictly subjected to the instructions of good manufacturing practice (GMP), which provides the standards for safety, efficacy and feasibility of therapeutic cells for large scale production [41]. Heterogeneity of hPSCs is reflected in the clonal variability of the cell lines, difference between cells within each clonal line during prolonged in vitro culture, as well as the continuous fluctuation of gene and protein expression levels at a frequency ranging from hours to days [34]. The hPSCs might undergo genetic mutations during long-term culture [34]. In addition, the differentiation of hPSCs in vitro can generate heterogenic subpopulation [34]. Therefore, development of effective and less labor-intensive approaches to regulate the purity and genetic background of hPSCs is important for product safety. Recently, the cost-effective scale-up in bioreactors has been used to overcome cumbersome labor-intensive production [42].
Low efficiency and reproducibility of differentiation
The differentiation of PSCs into desired cell types is a critical step in the development of PSC-based therapies. The predominant way to differentiate PSCs into various cell types is stepwise addition of specific growth factors, small molecules, and signaling inhibitors to mimic the corresponding developmental extracellular environment. However, there are significant challenges associated with the low efficiency and reproducibility of this process. Low efficiency can lead to the formation of cells that are functionally immature and not therapeutically relevant or that may even have tumorigenic potential, while lack of reproducibility can result in cell products that are variable in their potency and functionality. For example, many studies have reported that hESC-derived cardiomyocytes express marker TFs, show a cardiomyocyte-like phenotype, while display immature electrical phenotype [41]. Assurance of efficacy relies on a robust and reliable set of PSC differentiation protocols. Efforts to address these issues include the development of new culture conditions and genetic manipulation strategies that aim to improve cell differentiation and to ensure that the resulting cells are mature. Many groups have reported that modification of the timing, concentration and types of the growth factors, small molecules, signaling inhibitors can promote the efficiency and reproducibility of differentiation; while some other groups have reported that over-expression of master TFs can enhance cell differentiation [43], [44], [45], [46], [47]. In addition, efforts have been made to develop new culture platforms that allow for greater control over cell differentiation, including 3D bioreactors and microfluidics devices that mimic physiological environment [42]. Finally, a more effective solution is obtaining in-depth insight into the genetic regulatory networks governing stem cell differentiation in vivo and engineering them during in vitro differentiation. Analysis approaches combining gene expression data, proteomic pathway data, and cell signaling models have been created to explore key state transitions during differentiation [48].
Progress of PSC-derived multicellular systems
Generation of multicellular system to recapitulate physiological functions of tissue or organ in vitro is one of the most important and promising applications of PSCs for regenerative medicine. The basic rationale of multicellular system establishment is the capacity of self-assembling of cells which was discovered from reaggregation of a whole organism by sponge cells in 1907 [49]. Following this finding, more efforts have been paid to construct the complex tissue or organ by using dissociation-reaggregation approach in vitro. The breakthrough work of multicellular system generation was the successful establishment of 3D-mini-intestinal tissue with crypt-villus structures in vitro from single Lgr5+ stem cells in 2009 [50]. After that, several protocols have been developed to establish human organoids which are the micro-organ or tissue with physiological structure and functions in vitro, from either adult stem cells or PSCs, including generation of hPSC-derived intestinal organoids in 2011 [11], hPSC-derived optic cup organoids in 2012 [7], followed by other tissue types including cerebral [6], kidney [15], pancreas [13], and cardiac organoids [14] (Figure 2). To further increase the complexity of organoids in both cell-types and functions, researchers tried to mix different types of cells or organoids to form assembloids, such as the cortico-striatal assembloids with reflection of the interactive projections [51]. By assembling striatal organoids and cerebral cortical organoids, midbrain-microglia assembloids were generated with increased neuronal maturation and metabolic functions. Besides, other cutting-edge technologies have also been combined to form organoids with improved characteristics. Particularly, combination organoid technology with 3D-bioprinting has been widely used to establish vascularized organoids, such as gas-exchangeable lung organoids; or generate kidney organoids with high-homogeneity in high-throughput reported [52]. However, hPSC-derived organoids still have several key shortcomings hampering their application in regenerative medicine, including the challenges mentioned in the previous section: immune rejection; tumorigenicity; robust and homogeneous differentiation of specific cell types that can survive, proliferate and function maturely; inefficiency of scale-up and quality control for cell manufacture. Apart from these challenges, there are several additional key limitations in hPSC-based multicellular system generation, including disordered organ-specific morphological architecture, immature function, limited size and lifespan (Figure 2).
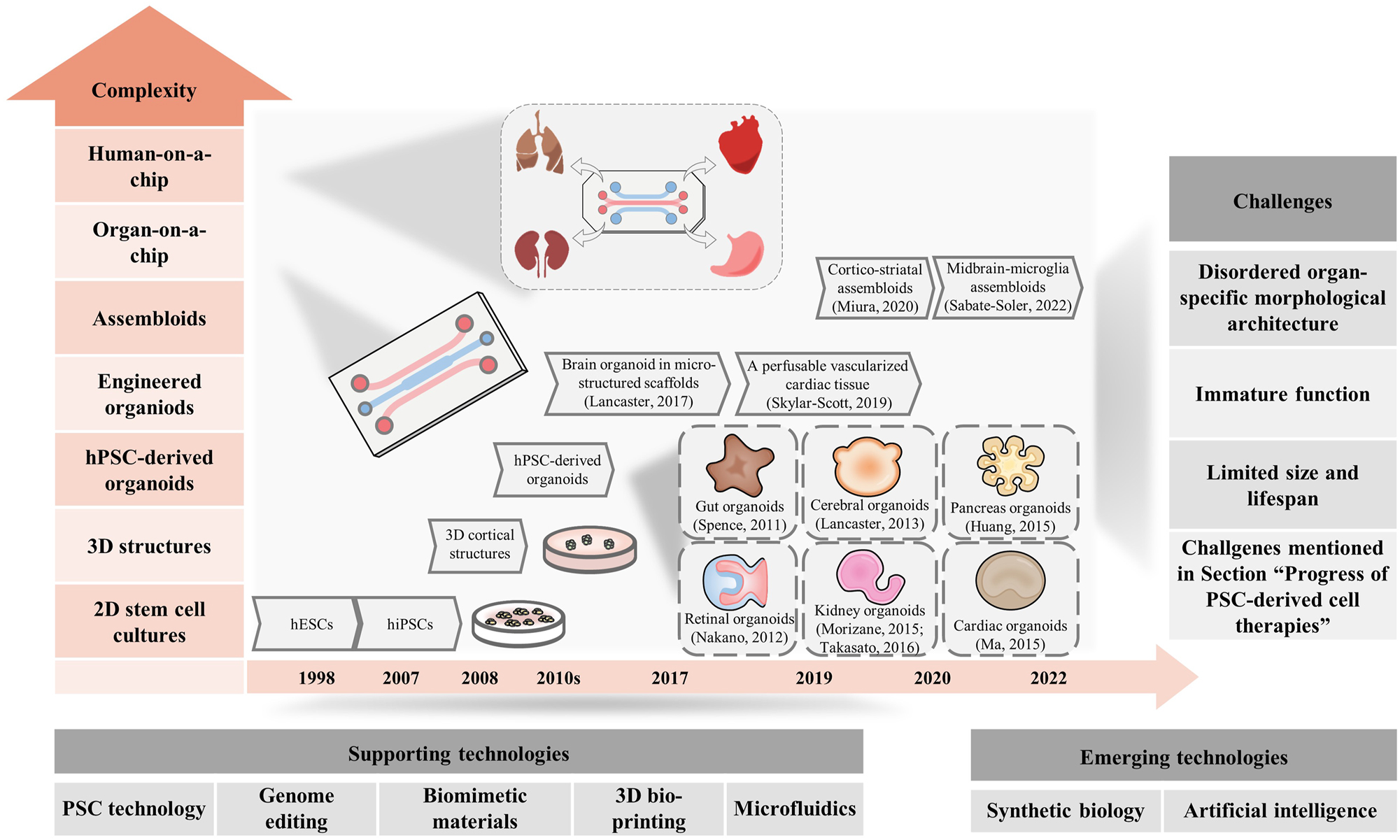
Progress and challenges of hPSC-derived organoids. The major progress of hPSC-derived organoids is shown, and the levels of complexity increases from bottom to up. The application of these innovative supporting technologies, combined with emergent technologies, are expected to address the challenges in this field.
Disordered organ-specific morphological architecture
Although organoids can display organized structure pattern compared with 2D monolayer cells, recapitulation of organ-specific spatiotemporal architecture still cannot be realized. Lack of necessary lineage cells like stroma cells and vascular endothelial cells is one reason for disordered structure. Another important reason is the absence of in vitro morphogen environment which plays a key role in developmental tissue organization in vivo. Providing morphogen gradient in vitro is promising to improve the structure of organoids, by either cell engineering, 3D-bioprinting, or other novel techniques.
Immature function
The predominant advantage of organoids is the improved tissue-like function filling the gap between 2D monolayer cell with limited function and the animal models. However, to date, majority of hPSC-derived organoids exhibit fetal cell-type composition and function analyzed by multi-omics, such as the liver organoids with fetal-like hepatocyte activity including cytochrome P459, vitamin A storage, lipid accumulation and inflammation response [53]; the cerebral cortical organoids resembling 19–24 post-conception-week prenatal brain [54]. Limited studies have reported to establish the PSC-derived organoids with neonatal function. In 2021, the establishment of hypothalamic arcuate organoids was reported with high similarity to neonatal human hypothalamus in cell type diversity and molecular signature, but whether possess the neonatal-level function was not demonstrated [55]. Optimization of differentiation protocols, integration of other lineages, re-construction of microenvironment might be the underlying strategy to overcome these challenges.
Limited size and lifespan
The limited lifespan and size of organoids are two related but different challenges in the field of organoid research. The limited lifespan and size are mainly caused by the limited nutrient supply and waste removal that occur as the organoid grows over time [56]. As organoids increase in size, the efficiency of nutrient supply and waste removal through diffusion decreases, which can give rise to limited growth, cell death and organoid necrosis. To address these challenges, researchers have developed various strategies such as using larger bioreactors [42] or using a perfusion system to improve nutrient access and waste removal [57]; improving vascularization to further spread nutrients through capillaries.
To realize organ or tissue replacement by PSC-derived organoids, development of innovative technologies, as well as combination with other disciplines are necessary to overcome some of the unsatisfactory outcomes of current organoids and further address the barriers.
Apart from the organoids, another type of multicellular system, called “synthetic embryos” including human blastoids has been established. Successful establishment of human blastoids were reported in 2021 by several groups, respectively [17], [18], [19], [20, 22]. Human blastoids were created via aggregation of ESCs, self-organization of naïve ESCs [17, 18], or even reprogramming of fibroblasts [20], respectively. Besides, another group generated human naïve PSC-derived blastoids which mimics the sequential lineage specification (trophectoderm, epiblast and primitive endoderm) of blastocyst development, and directionally attaches to endometrial cells with the polar trophectoderm to model implantation [58]. These synthetic embryos provide important multicellular models for developmental modeling, disease modeling, drug screening and organ regeneration in vitro.
To summarize, a major barrier to the clinical translation of PSC bioengineering efforts has been our achievement of a holistic understanding of the gene regulatory networks (GRNs) and biological processes governing cell behaviors inability to predictably and reproducibly control individual stem cell behaviors and their self-organization into multicellular systems. Stem cell engineering by synthetic biology can push the solutions.
Engineering PSCs with synthetic biology
Synthetic biology can be used to reinform or de novo design cellular behaviors from three standard modules: signal sensor (input), signal processing (process) and cellular responding (output) (Figure 3A). Generally, cells can sense diverse endogenous or artificial signals (input) by synthetic receptor or other sensing molecules, then they will undergo information processing through either endogenous regulatory networks or reinformed artificial genetic circuit (process), and finally produce presumptive designated cell behaviors (output). In synthetic biology toolkits, devices or circuits which can robustly and precisely sense and respond to diverse inputs (e.g., an extracellular signal) and result in customer-defined output (e.g., a transcription program) are important for cellular behaviors [29].
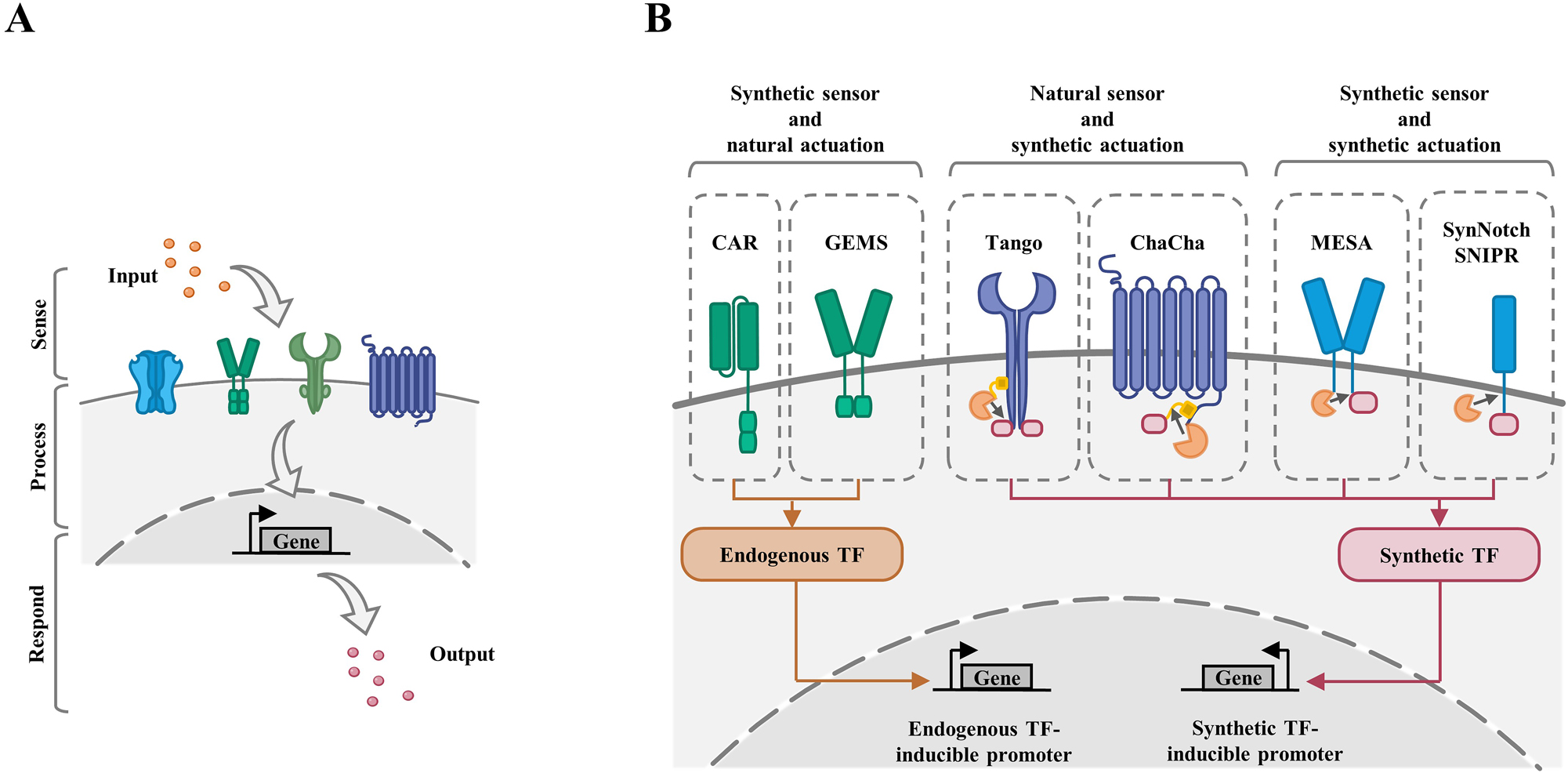
Principles and toolkit of synthetic receptors for programming cellular behaviors. (A) Cells can sense diverse endogenous or artificial signals (input) by sensors, then they will undergo information processing through either endogenous regulatory networks or reinformed artificial genetic circuits (process), and finally produce presumptive designated cell behaviors (output). (B) Mammalian synthetic receptors are categorized into three subtypes: synthetic sensor and natural actuation, natural sensor and synthetic actuation, and synthetic sensor and synthetic actuation. Examples of these subtypes are shown. CAR, chimeric antigen receptors; GEMS, generalized extracellular molecule sensor; MESA, modular extracellular signaling architecture; SynNotch, synthetic Notch; SNIPR, synthetic intramembrane proteolysis receptors.
The progress of programmable and modular receptor platforms allows researchers to rewire cellular input–output relationships. Synthetic receptors capable of novel input/output relationships can enable many applications, ranging from cell therapies to multicellular system formation [29].
Natural receptors are a class of proteins, which enable mammalian cells to recognize, process and respond to environmental information. Based on the understanding of natural receptors and the development of synthetic biology, scientists created synthetic receptors which can sense customized cues (input) and read out user-defined responses (output). Combing synthetic receptors with information processing tools, researchers can exert control over cellular behaviors, which has been well-summarized elsewhere [29]. To better design and expand the synthetic receptors, these receptors have been artificially divided into two modules: extracellular sensor and intracellular actuator. Briefly, the extracellular sensor is mainly to recognize the environmental signals, which can be either natural or artificial signals. In response to the input signal, the intracellular actuator will be stimulated and underwent either natural signal transduction or customized genetic regulation, executing certain functions such as apoptosis, proliferation, secretion and so forth. Sensor and actuator domain (natural or synthetic) can be modularly integrated (Figure 3B).
The most widely used receptors with synthetic sensor are chimeric antigen receptors (CARs), which rewire the T cell signaling pathways (TCRs) through replacing the native ligand-binding domains with antibody-based domains and retaining costimulatory receptor intracellular signaling domains artificially guiding T cells to robustly recognize and kill tumor cells with designated cell-surface antigens [59]. Receptors with synthetic sensor but natural actuation, including CAR [59] and generalized extracellular molecule sensor (GEMS) [41], allow to sense customized input and activate an endogenous genetic regulatory network to regulate cellular functions; receptors with natural sensor and synthetic actuation can execute customized behaviors or functions in response to sensor endogenous signal, such as the Tango [60] and ChaCha [61]; synthetic receptors with both synthetic sensors and synthetic actuators enable to better define both input and output, such as modular extracellular signaling architecture (MESA) [62], synthetic Notch receptor (synNotch) [63] and synthetic intramembrane proteolysis receptors (SNIPRs) [64] (Figure 3B). Transcriptional signal-processing is the downstream step linking actuators to gene transcription (output). By modifying or designing transcriptional regulatory systems, researchers can process signals transmitted downstream from natural or synthetic receptors (Figure 3B).
Programming PSCs with synthetic biology
Precise control of cell fate decision
One of the prominent advantages of hPSCs for cell therapy is the differentiation potency into various cell types, especially the cells which are hard to obtain from human tissues or possess limited proliferative capacity in vitro. In view of this, hPSCs have been regarded as a vital source to construct the pools of various types of cells by robust differentiation for cell therapy. However, based on current differentiation strategy using cocktail of small molecules and growth factor inhibitors, majority of hPSC-derived cells cannot be applied in clinical trials due to the immature function, low efficiency and high heterogeneity.
Direct differentiation of hPSCs into certain cell types or further organoids using forward programming approaches, including forced expression of TFs, provides a potential alternative. For example, thyroid follicular cells and transplantable organoids were differentiated from mouse and human PSCs through transient overexpression of NK2 homeobox 1 (NKX2-1) and paired box gene 8 (PAX8) [43, 44]. Besides, functional thyroid progenitors were robustly differentiated from mouse PSC-derived anterior foregut endoderm via transient and developmental stage-specific overexpression of NKX2-1 [47]. In addition, production of hPSC-derived megakaryocytes by overexpression of GATA1 (GATA binding protein 1), FLI1 (Fli-1 proto-oncogene, ETS transcription factor) and TAL1 (TAL bHLH transcription factor 1, erythroid differentiation factor) were reported as well [46]. Another group reported robust differentiating hPSCs into neurons, functional skeletal myocytes and oligodendrocytes [45].
However, such overexpression methods cannot precisely and robustly regulate cell fates because they fail in spatiotemporal controlling of gene expression and precisely controlling dosage of gene products. Precise encoding cell fate decision by synthetic lineage-controlling circuits holds great potential to obtain targeted cells with high efficiency and purity, facilitating their clinical application and scale-up production (Figure 4).
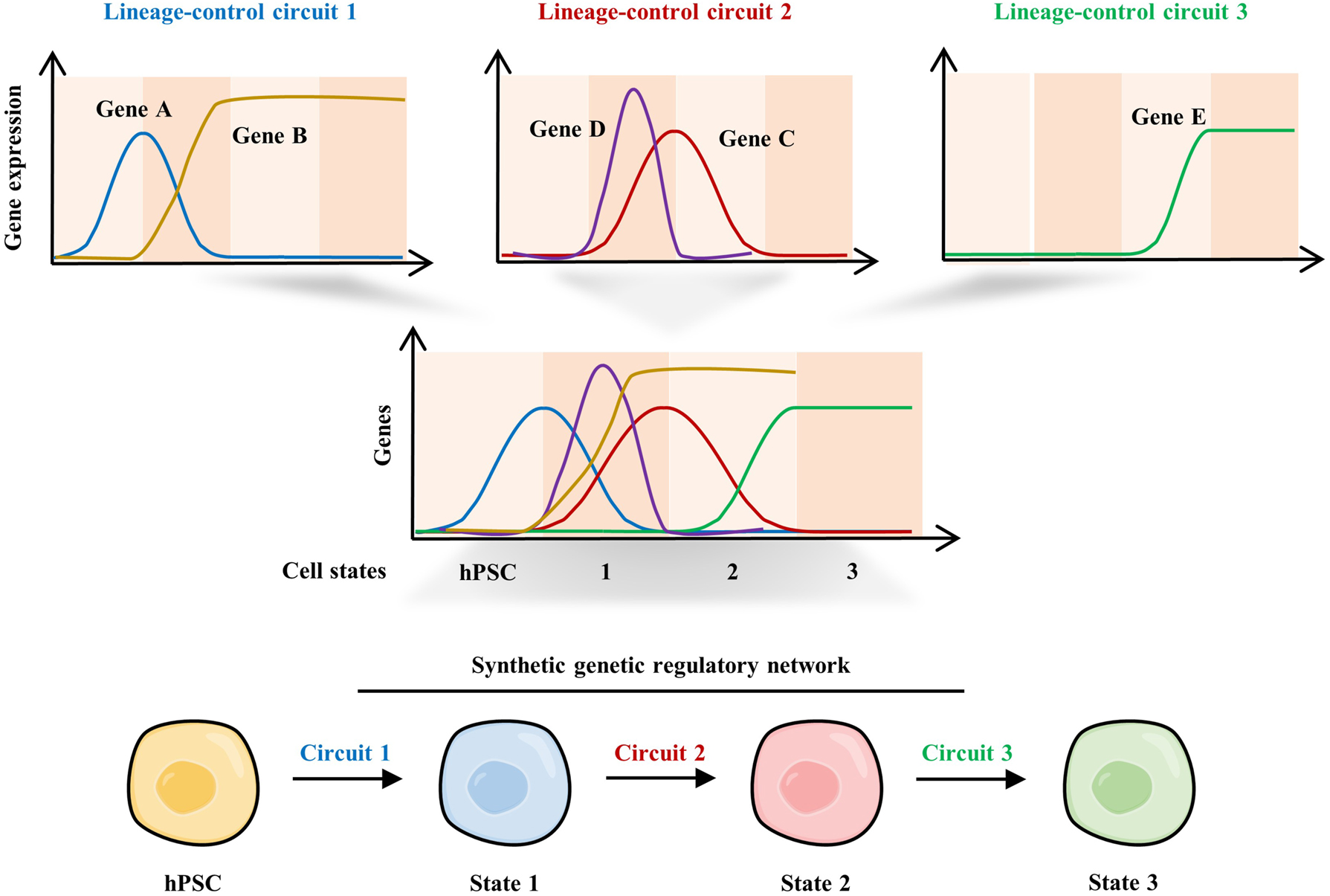
Control of PSC differentiation using complex synthetic lineage-control circuits. Schematic of synthetic genetic regulatory network programming differential expression dynamics of master transcription factors during hPSC differentiation. The three circuits provide programmable, mutually exclusive expression switches for gene A (ON-OFF-OFF), gene B (OFF-ON); gene C (OFF-ON-OFF), gene D (OFF-ON-OFF); and gene E (OFF-ON) expression in a temporal manner. These synthetic genetic circuits synergistically direct the differentiation of hPSCs towards specific cell types step by step.
A proof-of-concept work pointed out that synthetic genetic circuit would be a promising strategy to obtain hPSC-derived cells by directly controlling expression of master genes during development [65]. The design of lineage-control circuit was based on the previous studies of mouse pancreatic development, which demonstrated that the temporal expression pattern of neurogenin 3 (NGN3), pancreatic and duodenal homeobox 1 (PDX1) and V-maf musculoaponeurotic fibrosarcoma oncogene homolog A (MAFA) play a crucial role in the differentiation and maturation of pancreatic β cells. Thus, researchers designed a synthetic lineage-control network capable of mimicking the expression patterns of NGN3 (OFF-ON-OFF), PDX1 (ON-OFF-ON), and MAFA (OFF-ON) during in vivo pancreatic development by utilizing a vanillic acid dose-dependent signaling cascade and a gene switch. In hiPSC-derived pancreatic progenitor cells, this network enables correctly temporal differential expression of these three TFs and ultimately differentiates cells into glucose-sensitive insulin-secreting β-like cells. Notably, compared with cocktail-induced differentiation, these pancreatic β-like cells exhibited more similar dynamics of glucose-stimulated insulin release to that of human pancreatic islets, indicating the great potential of using synthetic gene circuits to establish the pool of functional cells for cell therapy (Figure 4).
Efficient purification of cell populations during PSC differentiation
The resulting heterogeneous population is a major problem of PSC differentiation. Thus, detection and purification of specific cell types for therapy is critical. Synthetic cell classifiers, usually based on microRNA (miRNA) switches, were created to detect and purify cells of interest. In 2015, Miki reported the establishment of synthetic miRNA switches to isolate desired cell type during hPSC differentiation [66]. Various miRNA switches were created, including miR-1,208a and 499a-5p for cardiomyocytes, miR-126 for endothelial cells, miR-122-5p for hepatocytes, and miR-375 for insulin-producing cells [66]. In detail, the miR-1 switch contains a synthetic mRNA encoding a fluorescent protein (FP) tagged with target site of miR-1, and desired cells expressing endogenous miR-1 will not express FP. And the miR-1-Bim switch, containing the apoptosis inducer Bim, will induce selective apoptosis in cells without miR-1 expression, namely non-target cells.
Enforcing enhanced or artificial therapeutic functions
Beyond achieving physiological functions, therapeutic function can be further enhanced, such as targeting and killing tumor cells. Predictable and defined levels of therapeutic functions must be achieved either by rewiring existing networks or engineering new ones. Synthetic biology is poised to enhance cell-based therapies by offering precise control over the context, intensity, and timing of therapeutic intervention. The therapeutic outputs of cells should sense inputs such as small molecules and disease biomarkers. After the signal processing, the dosage, timing, and localization of therapeutic gene expression or other therapeutic activities (outputs) should be tightly controlled.
A big milestone in this field was the FDA approval of Tisagenlecleucel, a CAR T cell product, in 2017 to treat refractory pre-B cell acute lymphoblastic leukemia and diffuse large B cell lymphoma. Specific immune cells including T cells and natural killer (NK) cells are key players of antitumor immunity, and the synthetic receptor CAR [54] can furthermore enhance their anti-tumor function. Such therapy consists of immune T cells taken from the patient, which are then engineered to express CAR that target cancer cells. After purification and expansion in vitro, the CAR T cells are injected into the patient. Although the therapeutic benefits of CAR-T cell therapy should not be overlooked, the limited cell sources, hard-to-be-engineered, time-consuming and money-consuming, heterogeneity of using primary immune cells hinder their applications in cancer therapy.
Combination of PSC differentiation technologies and synthetic CAR may address these challenges. Recently, one of the important examples is CAR iPSC-derived-NK cells [67]. Genetically modified iPSC-derived NK cells hold great potential to offer a safer, standardized and off-the-shelf antitumor therapy. Significantly, early-stage clinical trials of iPSC-derived-NK cells demonstrate safety and efficacy [67].
PSC-derived NK cells were initially differentiated from hESCs, and protocols have been modified to differentiate them from hiPSCs. Such engineered PSC-derived NK cells enable limitless and uniform NK cells derived from certain starting PSC line, and avoid collecting cells from individual donors or cord blood units. Besides, the starting PSC population can be more easily genetically modified than primary NK cells, especially when multiple genetic modifications (CAR and other modifications to enhance functions) are needed [68, 69], and the resulting NK cells derived from those engineered PSCs can express these genetic modifications more uniformly [70]. For example, in vivo persistence of cells is important for long-term therapeutic effects, and activation or inactivation of various genes (IL-15 overexpression or CISH knockout [71]) can enhance the expansion of certain cell types. However, such gene editing may disturb the differentiation process from PSCs to the final cell types (CISH in NK cells is a good example [71]), so in future inducible gene editing is required. PSCs engineered by genetic circuits containing CARs and licensed food additives-induced IL-15 overexpression and/or CISH knockout, may differentiate into NK cells with better persistence in vivo.
In addition, engineered cells with prostate-specific antigen (PSA)-specific GEMS were used to detect pathological PSA levels in the serum of patients with prostate cancer for diagnosis [72].
Dynamically monitoring and eliminating tumorigenic cells
One of the key concerns of hPSC application in cell therapy is the underlying tumorigenicity caused by undifferentiated PSCs and progenitor cells. Therefore, dynamically monitoring and effectively eliminating tumorigenic cells are critical to guarantee the safety of treatment. Engineering cells with inducible suicide or elimination switches can be a useful approach to simultaneously monitor and remove the risk of tumorigenesis.
Scientists reported that miRNA switches could sensitively identify and eliminate undifferentiated cells in iPSC-derived midbrain dopaminergic (mDA)-like neuronal cells [40]. As microRNA-302a-5p (miR-302a) is specifically expressed at high levels in PSCs, the group engineered a miR-302a switch in iPSCs, which functions to activate reporter protein translation when iPSCs differentiate into mDA cells with down-regulation of miR-302a. In SCID mice, miR-302a switch-sorted mDA cells mixed with iPSCs do not undergo teratoma formation. And when the puromycin-resistant gene is under the translational regulation of the miR-302a switch, puromycin-controlled selective elimination of iPSCs can be achieved in vitro.
The iPSC-derived neural stem/progenitor cells (iPSC-derived NS/PCs) are potential for the treatment of SCI, but they lead to tumorigenesis demonstrated by transplantation experiments in NOD/SCID mice. To address this issue, researchers reported the fail-safe systems that were designed based on suicide genes Caspase9 and HSVtk [38, 39]. By dosing NOD/SCID mice with a small molecule CID (AP20817) to induce Caspase9 expression, all engrafted cells in the injured spinal cord could rapidly orient to apoptosis [38]. Since ganciclovir (GCV) can be converted to cell cycle-dependent cytotoxic GCV-triphosphate by HSVtk, unlike the Caspase9/CID system, the HSVtk/GCV system can selectively ablate actively multiplying tumorigenic cells, including iPSCs and iPSC-derived NS/PCs [71]. Transplantation experiments in SCI mice model suggested that the HSVtk/GCV system prevented tumorigenic transformation of engrafted cells and that mature neural cells can preserve for maintaining the improvement of motor function.
In addition, our group also established a safety HLA-A11R hESC line immune-compatible with more than 20 % of Chinese population by installation of an AP1903-induced suicide switch [73]. AP1903 is the dimerizer agent rimiducid, and it will induce the caspase-9 based suicide system of the modified hESC and lead to cell death [73].
Together, engineering hPSCs with safety-guard circuits and therapeutic circuits might provide huge potential to broaden the application of hPSCs in cell therapy.
Applying synthetic biology approaches to construct PSC-derived multicellular systems
Engineering short-range cell-cell communication for self-organization
Synthetic biology can be applied to artificially construct direct or paracrine-mediated intercellular communication (Figure 5). Currently, the synNotch system is the most widely used synthetic receptor to manipulate the intercellular communication mediated by cell contact. This system contains a sender cell, whose cell surface expresses a specific antigen as a ligand; and a receiver cell, which expresses the corresponding synNotch receptor on the cell surface [63, 74]. The synNotch receptor is mainly composed of ECD, TMD and intracellular transcription factor domain. In view of the mechanism on signal transmission of the natural Notch pathway, when the sender cell contacts the receiver cell, the synNotch receptor of the receiver cell will recognize the corresponding antigen, and mediate the cleavage of the intracellular transcription factor domain, initiating the expression of target genes. The synNotch receptor system has various applications, such as achieving specific spatial arrangements and tracing differentiation of organoids. Firstly, by superimposing different synNotch receptor lines, a hierarchical structure of concentric circles [63] or concentric spheres [74] can be achieved in 2D or 3D. Secondly, this system was effectively used for lineage tracing or for reflecting the direct contact of cells during mouse embryonic development as well [75]. Third, engineering downstream responses of the synNotch receptor pathway can stimulate cell-fate decisions. This strategy was applied to mouse ESCs by engineering the synNotch receptor [76]. When Receiver cells are activated, the expression of the neuronal differentiation factor Neurogennin1 is initiated, thereby inducing the differentiation of receiver cells into neurons. Similarly, synNotch can be used to control other differentiation pathways, inducing the production of natural or synthetic morphogen [77], or inducing the expression of adhesion molecule [74] that leads to cell rearrangements.
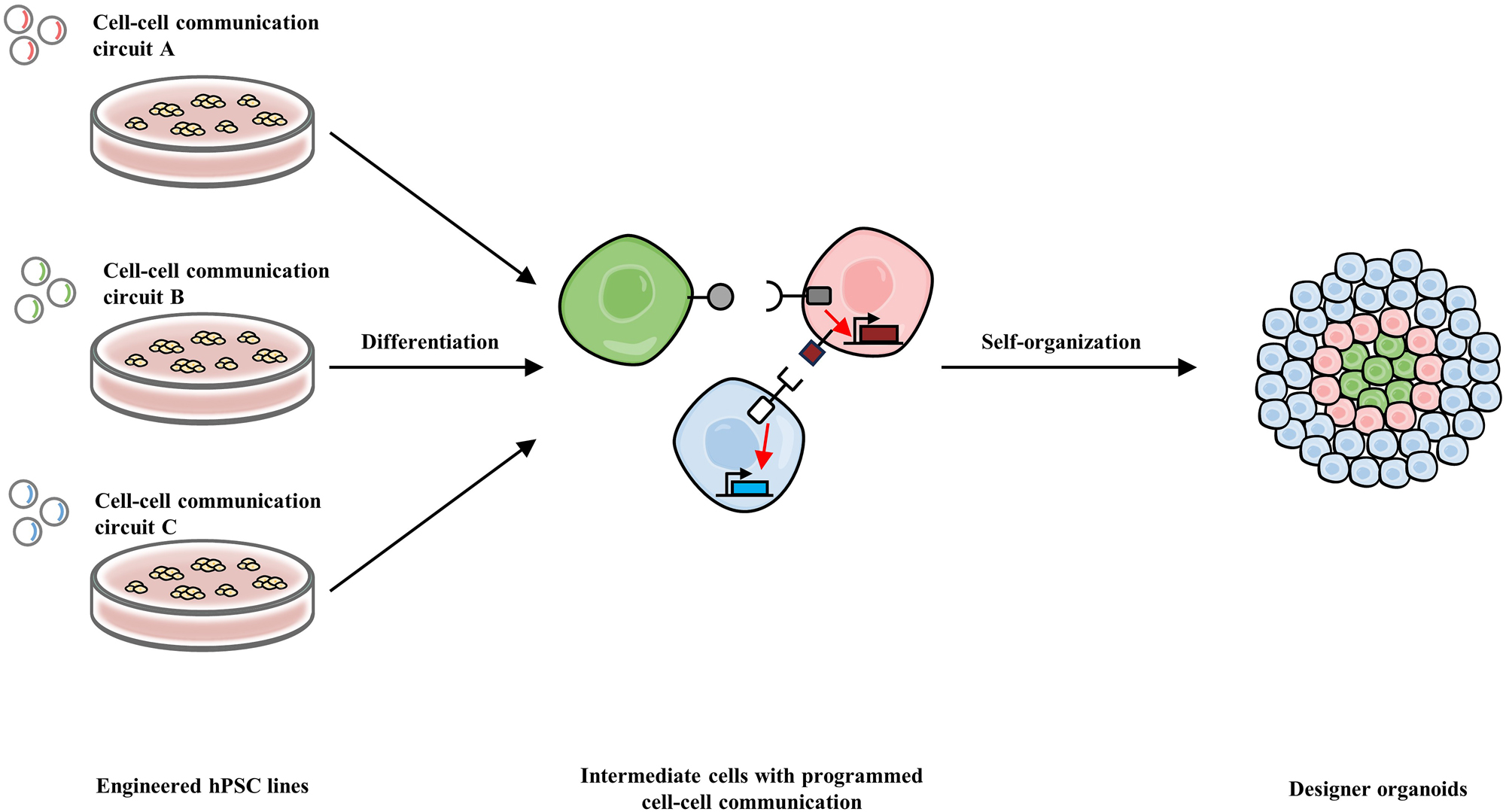
Manipulation of self-organization in hPSC-derived organoids via short-range cell-cell communication. Different synthetic cell-cell communication circuits (ligand-receptor pairs) are transferred into several hPSC lines, respectively. These engineered hPSC lines then undergo stepwise differentiation into certain progenitor cell types, respectively, which subsequently self-organize into designer organoids with specific spatial arrangement. The self-organization is directed by the ligand-receptor interaction, activation of receptors, and the activation of target genes including adhesion genes.
Recapitulating morphogen signaling to steer endogenous responses for multicellular system formation
In many developing organs, the spatiotemporal patterns of cellular position and fate decision are directed by graded concentrations of diffusible signal proteins, called morphogens. Cells dynamically regulate corresponding intrinsic GRNs in response to different concentrations of morphogens, obtain respective cell fates, and eventually form complex structure-organized multicellular system. In view of this, engineering stem cells to re-construct morphogen-mediated signaling or GRNs might facilitate the formation of multicellular system (Figure 6). Optogenetic chimeric receptors have been developed to manually control morphogen-mediated signaling or GRNs for mimicking the regulation of morphogen gradients. The basic principle to design optogenetic chimeric receptors is to fuse a photo-inducible domain to cytoplasmic domain of morphogen receptors, stimulating diverse morphogen signaling within mammalian cells, including bone morphogenetic protein (BMP), extracellular signal-regulated kinase (ERK), fibroblast growth factor (FGF), transforming growth factor beta (TGFβ) and Wnt [78], [79], [80]. Optogenetic chimeric receptor, which is constructed by fusing photolyase domain of blue-light photoreceptor Cryptochrome 2 (Cry2) to the cytoplasmic domain of canonical Wnt/β-catenin-response domain lipoprotein receptor-related protein 6 (LRP6), was used to mimic Wnt signaling within subpopulation of hESCs, facilitating hESC differentiation towards mesoderm with tissue-scale self-organized spatial patterns [78], indicating the great potential of generating multicellular system via directly engineering stem cells with synthetic receptors capable of mimicking morphogen signaling (Figure 6).
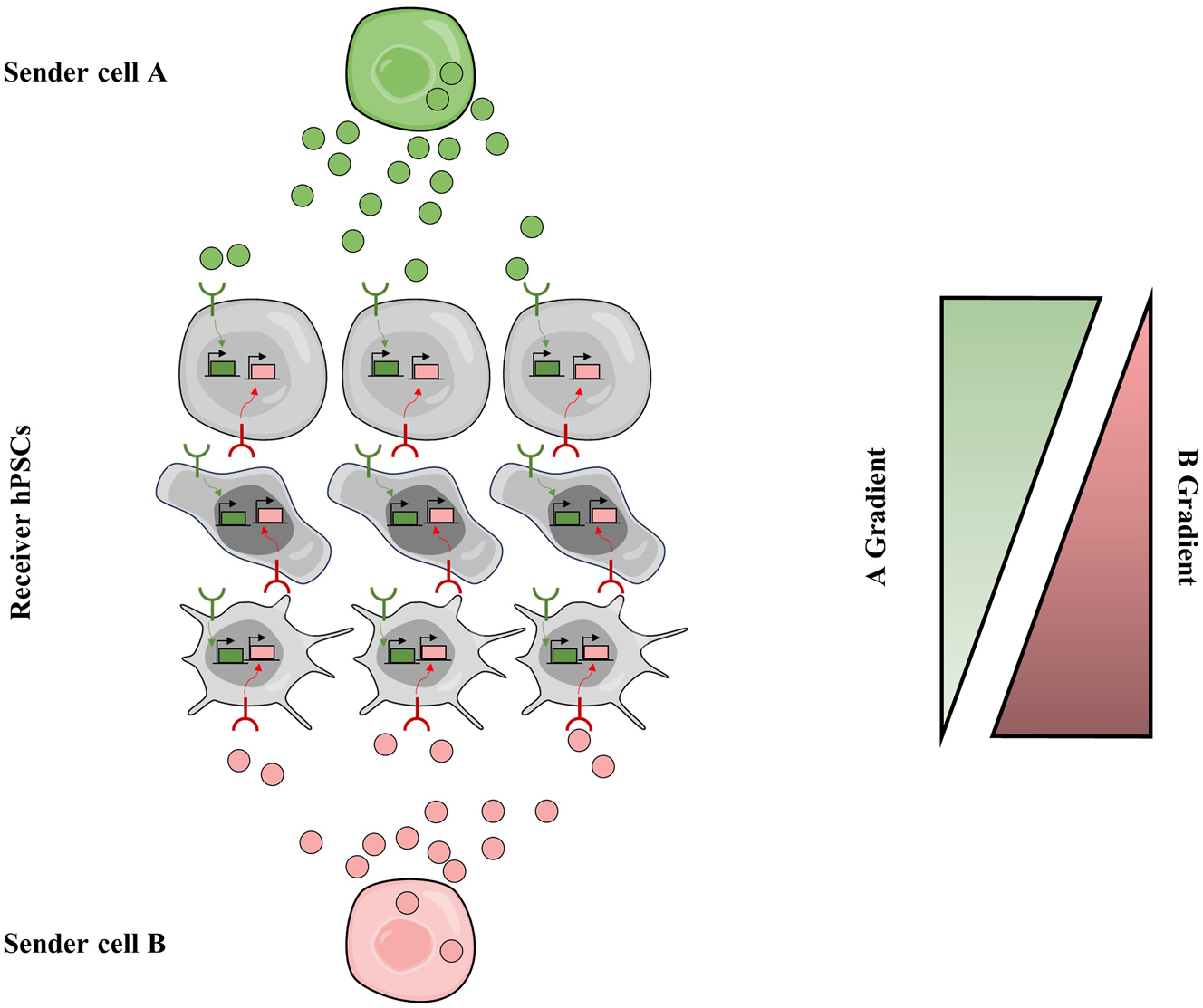
Recapitulation of artificial morphogen signaling via synthetic diffusible communication circuits. Type-A sender cell secret artificial morphogen A (green balls), type-B sender cells secret artificial morphogen B (red balls). Receiver hPSCs express both anti-A receptor and anti-B receptor, and the activation of receptors will trigger the expression of their target genes, respectively. Cells in the top sense strong A and weak B, cells in the middle sense moderate A and B, and those in the bottom sense strong B and weak A; and they differentiate into distinct cell types. Finally, multicellular patterning is programmed.
Apart from stimulating morphogen gradient-mediated signaling regulation, direct recapitulation of morphogen gradients could facilitate the construction of a multicellular system via precise control of spatial and temporal regulation. An orthogonal synthetic morphogen system has been developed by reforming synNotch system [77]. This morphogen system can sense soluble GFP secreted by secretor cells, and synergistically form GFP gradient by both anchor cells and anti-GFP receiver cells. Although this system was designed for orthogonal diffusible signal (GFP), GFP can be exchanged with any morphogen as long as a morphogen-specific anchor is available. With the increased number of synthetic receptors recognizing soluble factors, this proof-of-concept study paves the way for broader applications to generate multicellular systems.
Engineering cell-adhesion for multicellular architectures
As mentioned above, in vitro multicellular system formation mainly depends on self-assembling capacity, so high heterogeneity in both shape and cell-types might be due to the random cell self-aggregation instead of tight regulated cell-aggregation as in vivo. Therefore, artificial manipulation of cell-cell adhesion to define tissue-like structure might be another underlying approach to obtain multicellular system with high-fidelity architectures. To date, both natural and orthogonal cell-cell adhesion toolkits have been developed, including synNotch-cadherin, helixCAM and synCAM [74, 81], [82], [83]. Particularly, synNotch-cadherin system has been adapted and verified in mouse ESCs and chimeric mouse embryos [76], suggesting the applications of these cell-cell adhesion toolkits in various human PSC-derived cells for artificial assembling. Besides, these toolkits also exhibit great potential in regulating formation of embryoids, since engineering expression-levels of cell adhesion molecule has been reported to increase the assembling efficiency up to 3-fold of self-organized embryoid, which was formed by aggregation of ESCs, trophoblast stem cells and extraembryonic endoderm [84]. Together, harnessing these toolkits to manipulate structures of multicellular system could promote the self-aggregation efficiency and further obtain the designated organ-structures.
Precise control of multiple-lineage differentiation
Development process is tightly regulated by intracellular GRNs in response to extracellular signals or intracellular transduction. As mentioned above, current strategies to form organoids with multiple lineages are mainly by mixing of different lineages together or optimizing small-molecule cocktail to induce simultaneous differentiation of multiple lineages. However, the broader applications of these approaches are hindered, since cocktail protocol to induce arbitrarily combination of multiple-lineage differentiation, as well as culture medium to support arbitrarily multi-cell mixture are limited and difficult to develop. Recent years, the emerging of abundant omics data, particularly the single-cell transcriptomic profiles from diverse tissues and organs, presents an opportunity to compare gene expression patterns between hPSC-derived organoids and their in vivo counterparts, guiding the design of synthetic genetic circuits for controlling multiple-lineage differentiation and resulting in organoids with enhanced structures and function (Figure 7). One of the proof-of-concept studies demonstrated that human liver organoids with multi-lineages including hepatocyte, biliary, stellate cells and endothelial cells, were generated by using genetic circuits to up-regulate PROX1, ATF5 and CRISPR-based activation of endogenous CYP3A4, respectively [48].

Development of designer hPSC-derived organoids directed by synthetic genetic circuits. Comparison of expression profile between organoids and their native counterparts reveals several candidate TFs. Based on the TF candidates and their expression pattern, synthetic genetic circuits are designed, built, and transferred into hPSCs, which will direct the generation designer organoids with improved structures and functions. TFs, transcription factors.
Challenges and perspectives
The emergence of human PSCs fueled expectations for regenerative medicine including PSC-derived cell replacement therapy and establishment of PSC-derived multicellular systems. However, hPSC-based cell therapy is still far away from wide application in the clinic due to several major obstacles, including limited efficiency of current differentiation protocol of hPSC towards both specific types of cells or organoids; the immunogenicity of hESCs for allogeneic treatment of either cell therapy or further organ transplantation; and the underlying tumorigenicity of hPSCs as well as hPSC-derived progenitor cells. As reviewed above, engineering PSCs with synthetic biology tools provides creative strategies to address these problems (Figure 8). Furthermore, limitations and perspectives of this field will be discussed below.
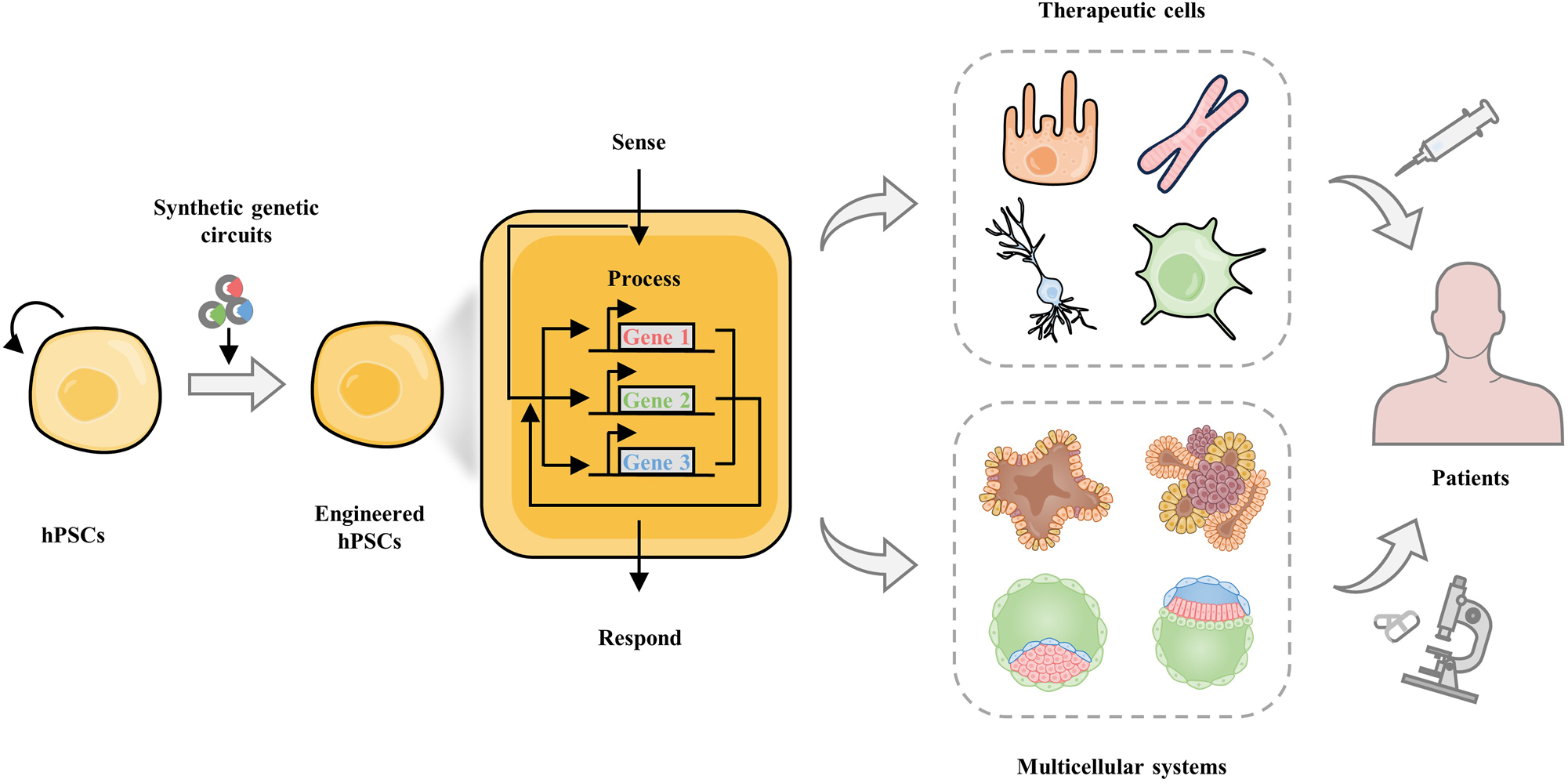
Engineering hPSCs by synthetic biology for regenerative medicine. hPSCs have exhibited prominent applications in regenerative medicine, including hPSC-based cell therapy and hPSC-derived multicellular systems for organ transplantation in future. Engineered hPSCs with synthetic genetic circuits can drive novel sense-process-respond in hPSCs and their derivatives, and facilitate the establishment of hPSC-derived therapeutic cells and multicellular systems.
Investigation and extraction of GRNs of cell state transitions bolsters the design of lineage-controlling and state-switch circuits
To precisely control cell fate determination or cell state, a clear view of GRNs of different cell types is necessary, since these GRNs will provide critical cues for stem cell engineering. However, majority cell type-specific GRNs for cell fate determination and function reconstruction are largely unclear. With the fast development of system biology and artificial intelligence (AI) technology, single or multigene perturbation model have been developed to extract and predict the impacts of genes on transcriptional outcomes. Therefore, integration of this data-driven science into stem cell engineering will provide more comprehensive and novel information about gene functions or GRNs, providing the basic cues and strategies for stem cell engineering. Omics data can be used to analyze and understand the cell state transition process. This can be achieved through systematic experiments and/or by utilizing AI models. We may be able to figure out the key GRNs and key TFs that regulate this process. For instance, based on a library of ORFs including different splicing forms of all human TFs, a TF atlas of expression profiles was conducted by overexpressing individual TFs in ESCs in 2023 [85]. They provided a systematic platform for TF screening, contributing to our comprehensive understanding of GRNs. Besides, benefiting from the application of machine learning in biology, such as cSTAR [86], we can use computational models to integrate different omics data to predict mechanistic networks during cell state transitions. These new insights will facilitate our purposeful design of genetic circuits to precisely manipulate and control the fate of stem cells.
Deciphering the biological regulatory mechanisms of self-organizing events during development facilitates engineering multicellular systems using synthetic biology
Beyond revealing the basic principles that govern individual PSC behaviors, we still need to understand in-depth how the collective behaviors of cells give rise to multicellular behaviors and self-organizing events such as pattern formation and symmetry breaking. In detail, we require to investigate how to spatially arrange diverse cell types and how to establish boundary conditions during in vitro culture, including extracellular matrix (ECM) composition and organ geometry.
Creating approaches controlling microenvironment including ECM for better multicellular systems
So far, several synthetic genetic circuits controlling input, processing, and output mechanisms have been engineered. However, there are still some developmental participants for which there is a lack of synthetic biology approaches to control them. For instance, the ECM is an important one for signaling transduction among cells. After produced and modified by sender cells, ECM presents signals to receiver cells through its molecular components and mechanical properties. Nevertheless, due to their large size and complex assembly, we can hardly regulate the production, modification, and signal transduction of ECM, which is important for the development of organoids. Recently, machine-learning algorithm, ProteinMPNN was used to design many novel fibrous proteins [87]. The mechanical properties of the artificial fibers can be modulated by changing the pore size of the fibers, in which smaller pore sizes result in stiffer fibers. Such tunability of the fibers will be useful for protein-based hydrogels (a kind of ECM), where the bulk moduli can be rationally changed by using fibers of different porosity with applications in tissue engineering.
Engineering “designer” PSCs or their derivatives with closed-loop circuit to monitor and cure diseases dynamically
Synthetic biology tools offer a unique opportunity to engineer “designer” stem cells with closed-loop circuits for monitoring and dynamically treating diseases. “Designer” stem cells are precisely engineered to sense and respond to their environment, secreting specific molecules to regulate cell fate, function, and behavior. By combining stem cells with closed-loop circuits, it may be possible to achieve an enhanced level of control over therapeutic interventions, personalizing treatment based on real-time disease states. One potential application of this technology is in monitoring and treating cancer. “Designer” PSC-derived immune cells can be engineered to express receptors that bind to specific cancer biomarkers, triggering a signaling cascade that leads to the secretion of therapeutic agents. These agents can include cytokines, chemokines, or small molecule drugs that specifically target cancer cells while minimizing side effects on surrounding tissues. Another potential application is in monitoring and treating diabetes. Researchers have established β-cell-mimetic “designer” cells via engineering HEK-293 cells with closed-loop circuit coupling glycolysis-mediated calcium entry to an excitation-transcription system which activates insulin or GLP-1 expression [88]. This circuit is capable of sensing glucose levels dynamically and resulting in secreting insulin or other regulatory molecules. Besides, this circuit can be optimized to secrete growth factors or cytokines as well in order to promote pancreatic cell regeneration or regulate immune responses to improve glycemic control. In the future, “designer” PSC-derived β cells engineered by such optimized closed-loop circuit may be used to monitor and treat diabetes.
Generating better PSC-derived cellular or multicellular models for drug discovery by combining synthetic biology with AI
Synthetic gene circuits, which enable reporting and tracking disease-associated molecular events, hold great potential to improve the precision and speed in phenotype-based drug discovery (PDD) based on assays in cells and complex cellular systems, including hPSC-derived- cells, organoids, and microphysiological systems (MPS) such as organ-on-a-chip [89]. However, one important challenge is limited predictable de novo design of circuits. The using of AI may enable automated design of robust gene circuits that can monitor desired pathways and phenotypes. In the future, for example, by engineering hPSC-derived neurons using genetic circuits, these neurons may be capable of identifying and tracking systems-wide, neurodegenerative-disease-associated phenotypes, including aggregate formation based on a complex reporter circuit [89] and hyperexcitability by another frequency-decoding circuit [89], which will largely facilitate drug discovery.
Promoting the specificity, orthogonality, versatility of synthetic biology tools
Synthetic biology, with its promise to revolutionize stem cell engineering, is actively seeking to promote the specificity, orthogonality, and versatility of its tools. Different from the genetic network of bacteria and yeast, GRNs in mammalian cells, including hPSCs are pretty complex. The specificity of synthetic biology tools lies in their ability to target and modulate specific biological processes without affecting others. This ability is achieved through the use of specific ligands, TFs, or miRNAs, which merely bind to their targets. Orthogonality means that different gene circuits can interact with each other accurately without causing cross-talk or side effects. To achieve high orthogonality, precise and detailed regulation and optimization of each component in the circuits are required. First, orthogonal genetic operators need to be used to ensure their spatial and temporal independence. Second, orthogonal regulatory elements should be selected to construct interactive networks between different gene circuits, enabling each part to respond independently to different inputs. Third, using appropriate expression vectors and cell systems is crucial as well, because they can affect the expression level of gene circuits and the viability of cells. Besides, the versatility of synthetic circuits to engineer different cell types during the differentiation of PSCs is difficult because most genetic components, especially those that rely on transcriptional regulation, perform very differently in different cell types due to incomplete isolation from the endogenous GRNs.
In addition, cellular burden imposed by genetic circuits engineering in mammalian cells has also been reported. Artificial reconstruction of cellular functions results in unintended dynamic coupling of exogenous genetic circuits with endogenous gene expression due to limited cellular resources. This cellular burden could be attenuate by co-expressing with microRNAs and RNA binding proteins, indicating a more rational design of genetic circuits and optimized engineering strategy might be a promising way to maximum actuation of cell functions, balancing the cellular burden and functional outputs [90].
Creating novel synthetic biology toolkits and modules for PSC engineering
These toolkits are mostly suitable for bacteria and yeast, facilitating the behaviors control or metabolic network engineering. Unfortunately, most of these toolkits do not work in mammalian cells, indicating that establishment of molecule element toolkits for stem cell engineering is necessary. Construction of both natural molecule-based toolkits and de novo design of artificial DNA sequences or protein domains are the approaches that could be used. Recently, a study has harnessed the power of synthetic biology to significantly extend yeast cellular lifespan. The authors successfully rewired a natural toggle switch in yeast cells to create a genetic clock (based on lysine deacetylase Sir2 and heme-activated protein (HAP)) that oscillates between nucleolar and mitochondrial aging processes, which can effectively delay cellular aging. This research may set the stage for future applications in regenerative medicine and the treatment of age-related diseases [91]. In the future, such an application would require additional research and development in human cells. As for synthetic multicellular systems, novel available tools may be used to realize diverse developmental processes. For example, the development of synthetic TFs, multi-input logic gates, and other synthetic genetic elements is beneficial to design a simple, universal and expandable lineage-control network, and even realize the goal of multiple fates for one cell. Recently, MultiFate, a synthetic gene circuit, was reported to programme many stable states in one mammalian cell [92]. This system uses programmable zinc finger TFs which can self-activate as homodimers and mutually inhibit one another when forming heterodimers. Such homodimerization or heterodimerization is controlled by small molecules. As a result, by expressing three types of these TFs, one cell line can generate seven different cell states. In the future, MultiFate can be combined with other synthetic circuits mentioned above and enable stem cells to make a series of fate choices during the formation of multicellular systems. Besides, this field will not merely be restricted to the creation of new synthetic biology tools. Combinations of different tools to control different developmental processes, such as cell sorting and pattern formation, will be used to develop more complex organoids and synthetic embryos in vitro. Furthermore, with more available tools, researchers will be able to study the interplay of developmental processes in multicellular systems. For example, the use of optogenetic tools to manipulate tissue shape represents the opportunity to alter organoid shape and study the feedback of shape change on other aspects of development.
Directly engineering stem cell to regenerate human tissues in situ
The final goal of regenerative medicine is to restore the functions of the disabled body in vivo. Current strategies mainly focus on engineering cells in vitro, followed by reinjection into human body, such as CAR-T therapy. Although these approaches have exhibited dramatic improvement of patient survival, engineering cells in vitro requires high-quality operation and environment to ensure the safety of therapy, increasing the risks of immunogenicity and the costs of treatment. In vivo delivery of genetic circuits into stem cells will significantly reduce these hurdles. To fulfill this goal, delivery vectors with high delivering efficiency, specific tropism and low immunogenicity are important. To date, the major in vivo gene delivery system uses viral vectors such as AAVs and their modified variants and non-viral vectors such as LNPs, EVs and VLPs [93], [94], [95]. Though several pre-clinical and clinical trials have already achieved great success using these tools for in vivo treatments [96, 97], to further realize the delivery of genetic circuits for in situ regeneration of injured or dysregulated tissues, more attention needs to be paid to the development of gene delivery technologies.
Improving the research regulations of engineered hPSCs
In some countries or regions, like China, the USA and Europe, relevant guidelines have been issued to put forward special considerations and requirements for genetically modified cellular therapy products. Genetically engineered PSC-based cell therapies will carry new security risks, especially those based on over-engineered PSCs loaded complicated synthetic genetic circuits. In the future, governments from different countries and regions may need to adjust research policies and regulatory approaches, and strive to adopt more internationally recognized principles and rules [98].
Funding source: National Key Research and Development Program
Award Identifier / Grant number: (2019YFA0903800, 2019YFA0110800 to W.L., 2022YFA0806302 to S.W.)
Funding source: National Natural Science Foundation of China
Award Identifier / Grant number: (32225030 to W.L.)
Funding source: CAS Project for Young Scientists in Basic Research
Award Identifier / Grant number: (YSBR-012 to W.L.)
-
Research ethics: Not applicable.
-
Informed consent: Not applicable.
-
Author contributions: Yihuan Mao, Siqi Wang: Conceptualization, Investigation, Writing-Original Draft, Writing – Review & Editing. Jiazhen Yu: Investigation, Writing-Original Draft, Writing – Review & Editing. Wei Li: Conceptualization, Writing – Review & Editing, Supervision, Funding acquisition. The authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Competing interests: The authors state no conflict of interest.
-
Research funding: This study was supported by grants from the National Key Research and Development Program (2019YFA0903800, 2019YFA0110800 to W.L., 2022YFA0806302 to S.W.), the National Natural Science Foundation of China (32225030 to W.L.), the CAS Project for Young Scientists in Basic Research (YSBR-012 to W.L.).
-
Data availability: Not applicable.
References
1. Tewary, M, Shakiba, N, Zandstra, PW. Stem cell bioengineering: building from stem cell biology. Nat Rev Genet 2018;19:595–614. https://doi.org/10.1038/s41576-018-0040-z.Search in Google Scholar PubMed
2. Thomson, JA, Itskovitz-Eldor, J, Shapiro, SS, Waknitz, MA, Swiergiel, JJ, Marshall, VS, et al.. Embryonic stem cell lines derived from human blastocysts. Science 1998;282:1145–7. https://doi.org/10.1126/science.282.5391.1145.Search in Google Scholar PubMed
3. Takahashi, K, Tanabe, K, Ohnuki, M, Narita, M, Ichisaka, T, Tomoda, K, et al.. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–72. https://doi.org/10.1016/j.cell.2007.11.019.Search in Google Scholar PubMed
4. Yu, J, Vodyanik, MA, Smuga-Otto, K, Antosiewicz-Bourget, J, Frane, JL, Tian, S, et al.. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917–20. https://doi.org/10.1126/science.1151526.Search in Google Scholar PubMed
5. Storb, R. Edward Donnall Thomas (1920–2012). Nature 2012;491:334. https://doi.org/10.1038/491334a.Search in Google Scholar PubMed
6. Lancaster, MA, Renner, M, Martin, CA, Wenzel, D, Bicknell, LS, Hurles, ME, et al.. Cerebral organoids model human brain development and microcephaly. Nature 2013;501:373–9. https://doi.org/10.1038/nature12517.Search in Google Scholar PubMed PubMed Central
7. Nakano, T, Ando, S, Takata, N, Kawada, M, Muguruma, K, Sekiguchi, K, et al.. Self-Formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012;10:771–85. https://doi.org/10.1016/j.stem.2012.05.009.Search in Google Scholar PubMed
8. Dye, BR, Hill, DR, Ferguson, MA, Tsai, YH, Nagy, MS, Dyal, R, et al.. In vitro generation of human pluripotent stem cell derived lung organoids. Elife 2015;4:e05098. https://doi.org/10.7554/elife.05098.Search in Google Scholar
9. McCracken, KW, Catá, EM, Crawford, CM, Sinagoga, KL, Schumacher, M, Rockich, BE, et al.. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014;516:400–4. https://doi.org/10.1038/nature13863.Search in Google Scholar PubMed PubMed Central
10. Múnera, JO, Sundaram, N, Rankin, SA, Hill, D, Watson, C, Mahe, M, et al.. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell 2017;21:51–64.e6. https://doi.org/10.1016/j.stem.2017.05.020.Search in Google Scholar PubMed PubMed Central
11. Spence, JR, Mayhew, CN, Rankin, SA, Kuhar, MF, Vallance, JE, Tolle, K, et al.. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011;470:105–9. https://doi.org/10.1038/nature09691.Search in Google Scholar PubMed PubMed Central
12. Takebe, T, Sekine, K, Enomura, M, Koike, H, Kimura, M, Ogaeri, T, et al.. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013;499:481–4. https://doi.org/10.1038/nature12271.Search in Google Scholar PubMed
13. Huang, L, Holtzinger, A, Jagan, I, BeGora, M, Lohse, I, Ngai, N, et al.. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med 2015;21:1364–71. https://doi.org/10.1038/nm.3973.Search in Google Scholar PubMed PubMed Central
14. Ma, Z, Wang, J, Loskill, P, Huebsch, N, Koo, S, Svedlund, FL, et al.. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat Commun 2015;6:7413. https://doi.org/10.1038/ncomms8413.Search in Google Scholar PubMed PubMed Central
15. Takasato, M, Er, PX, Chiu, HS, Maier, B, Baillie, GJ, Ferguson, C, et al.. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 2015;526:564–8. https://doi.org/10.1038/nature15695.Search in Google Scholar PubMed
16. Wimmer, RA, Leopoldi, A, Aichinger, M, Wick, N, Hantusch, B, Novatchkova, M, et al.. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 2019;565:505–10. https://doi.org/10.1038/s41586-018-0858-8.Search in Google Scholar PubMed PubMed Central
17. Yu, L, Wei, Y, Duan, J, Schmitz, DA, Sakurai, M, Wang, L, et al.. Blastocyst-like structures generated from human pluripotent stem cells. Nature 2021;591:620–6. https://doi.org/10.1038/s41586-021-03356-y.Search in Google Scholar PubMed
18. Yanagida, A, Spindlow, D, Nichols, J, Dattani, A, Smith, A, Guo, G. Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell 2021;28:1016–22 e4. https://doi.org/10.1016/j.stem.2021.04.031.Search in Google Scholar PubMed PubMed Central
19. Sozen, B, Jorgensen, V, Weatherbee, BAT, Chen, S, Zhu, M, Zernicka-Goetz, M. Reconstructing aspects of human embryogenesis with pluripotent stem cells. Nat Commun 2021;12:5550. https://doi.org/10.1038/s41467-021-25853-4.Search in Google Scholar PubMed PubMed Central
20. Liu, X, Tan, JP, Schroder, J, Aberkane, A, Ouyang, JF, Mohenska, M, et al.. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 2021;591:627–32. https://doi.org/10.1038/s41586-021-03372-y.Search in Google Scholar PubMed
21. Heidari Khoei, H, Javali, A, Kagawa, H, Sommer, TM, Sestini, G, David, L, et al.. Generating human blastoids modeling blastocyst-stage embryos and implantation. Nat Protoc 2023;18:1584–620. https://doi.org/10.1038/s41596-023-00802-1.Search in Google Scholar PubMed
22. Fan, Y, Min, Z, Alsolami, S, Ma, Z, Zhang, E, Chen, W, et al.. Generation of human blastocyst-like structures from pluripotent stem cells. Cell Discov 2021;7:81. https://doi.org/10.1038/s41421-021-00316-8.Search in Google Scholar PubMed PubMed Central
23. Amadei, G, Handford, CE, Qiu, C, De Jonghe, J, Greenfeld, H, Tran, M, et al.. Embryos complete gastrulation to neurulation and organogenesis. Nature 2022;610:143–53. https://doi.org/10.1038/s41586-022-05246-3.Search in Google Scholar PubMed PubMed Central
24. Gardner, TS, Cantor, CR, Collins, JJ. Construction of a genetic toggle switch in Escherichia coli. Nature 2000;403:339–42. https://doi.org/10.1038/35002131.Search in Google Scholar PubMed
25. Elowitz, MB, Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000;403:335–8. https://doi.org/10.1038/35002125.Search in Google Scholar PubMed
26. Guet, CC, Elowitz, MB, Hsing, W, Leibler, S. Combinatorial synthesis of genetic networks. Science 2002;296:1466–70. https://doi.org/10.1126/science.1067407.Search in Google Scholar PubMed
27. Basu, S, Gerchman, Y, Collins, CH, Arnold, FH, Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 2005;434:1130–4. https://doi.org/10.1038/nature03461.Search in Google Scholar PubMed
28. Friedland, AE, Lu, TK, Wang, X, Shi, D, Church, G, Collins, JJ. Synthetic gene networks that count. Science 2009;324:1199–202. https://doi.org/10.1126/science.1172005.Search in Google Scholar PubMed PubMed Central
29. Manhas, J, Edelstein, HI, Leonard, JN, Morsut, L. The evolution of synthetic receptor systems. Nat Chem Biol 2022;18:244–55. https://doi.org/10.1038/s41589-021-00926-z.Search in Google Scholar PubMed PubMed Central
30. Schwartz, SD, Regillo, CD, Lam, BL, Eliott, D, Rosenfeld, PJ, Gregori, NZ, et al.. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet 2015;385:509–16. https://doi.org/10.1016/s0140-6736(14)61376-3.Search in Google Scholar
31. Kamao, H, Mandai, M, Okamoto, S, Sakai, N, Suga, A, Sugita, S, et al.. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep 2014;2:205–18. https://doi.org/10.1016/j.stemcr.2013.12.007.Search in Google Scholar PubMed PubMed Central
32. Menasché, P, Vanneaux, V, Hagège, A, Bel, A, Cholley, B, Parouchev, A, et al.. Transplantation of human embryonic stem cell–derived cardiovascular progenitors for severe ischemic left ventricular dysfunction. J Am Coll Cardiol 2018;71:429–38. https://doi.org/10.1016/j.jacc.2017.11.047.Search in Google Scholar PubMed
33. Wang, YK, Zhu, WW, Wu, MH, Wu, YH, Liu, ZX, Liang, LM, et al.. Human clinical-grade parthenogenetic ESC-derived dopaminergic neurons recover locomotive defects of nonhuman primate models of Parkinson’s disease. Stem Cell Rep 2018;11:171–82. https://doi.org/10.1016/j.stemcr.2018.05.010.Search in Google Scholar PubMed PubMed Central
34. Yamanaka, S. Pluripotent stem cell-based cell therapy—promise and challenges. Cell Stem Cell 2020;27:523–31. https://doi.org/10.1016/j.stem.2020.09.014.Search in Google Scholar PubMed
35. Deuse, T, Hu, X, Gravina, A, Wang, D, Tediashvili, G, De, C, et al.. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat Biotechnol 2019;37:252–8. https://doi.org/10.1038/s41587-019-0016-3.Search in Google Scholar PubMed PubMed Central
36. Deuse, T, Seifert, M, Phillips, N, Fire, A, Tyan, D, Kay, M, et al.. Human leukocyte antigen I knockdown human embryonic stem cells induce host ignorance and achieve prolonged xenogeneic survival. Circulation 2011;124:S3–9. https://doi.org/10.1161/circulationaha.111.020727.Search in Google Scholar
37. Lezmi, E, Jung, J, Benvenisty, N. High prevalence of acquired cancer-related mutations in 146 human pluripotent stem cell lines and their differentiated derivatives. Nat Biotechnol 2024. https://doi.org/10.1038/s41587-023-02090-2 [Epub ahead of print]. Search in Google Scholar PubMed
38. Itakura, G, Kawabata, S, Ando, M, Nishiyama, Y, Sugai, K, Ozaki, M, et al.. Fail-safe system against potential tumorigenicity after transplantation of iPSC derivatives. Stem Cell Rep 2017;8:673–84. https://doi.org/10.1016/j.stemcr.2017.02.003.Search in Google Scholar PubMed PubMed Central
39. Kojima, K, Miyoshi, H, Nagoshi, N, Kohyama, J, Itakura, G, Kawabata, S, et al.. Selective ablation of tumorigenic cells following human induced pluripotent stem cell-derived neural stem/progenitor cell transplantation in spinal cord injury. Stem Cells Transl Med 2019;8:260–70. https://doi.org/10.1002/sctm.18-0096.Search in Google Scholar PubMed PubMed Central
40. Parr, CJ, Katayama, S, Miki, K, Kuang, Y, Yoshida, Y, Morizane, A, et al.. MicroRNA-302 switch to identify and eliminate undifferentiated human pluripotent stem cells. Sci Rep 2016;6:32532. https://doi.org/10.1038/srep32532.Search in Google Scholar PubMed PubMed Central
41. Blau, HM, Daley, GQ. Stem cells in the treatment of disease. N Engl J Med 2019;380:1748–60. https://doi.org/10.1056/nejmra1716145.Search in Google Scholar
42. Qian, X, Jacob, F, Song, MM, Nguyen, HN, Song, H, Ming, GL. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat Protoc 2018;13:565–80. https://doi.org/10.1038/nprot.2017.152.Search in Google Scholar PubMed PubMed Central
43. Romitti, M, Tourneur, A, de Faria da Fonseca, B, Doumont, G, Gillotay, P, Liao, XH, et al.. Transplantable human thyroid organoids generated from embryonic stem cells to rescue hypothyroidism. Nat Commun 2022;13:7057. https://doi.org/10.1038/s41467-022-34776-7.Search in Google Scholar PubMed PubMed Central
44. Antonica, F, Kasprzyk, DF, Opitz, R, Iacovino, M, Liao, XH, Dumitrescu, AM, et al.. Generation of functional thyroid from embryonic stem cells. Nature 2012;491:66–71. https://doi.org/10.1038/nature11525.Search in Google Scholar PubMed PubMed Central
45. Pawlowski, M, Ortmann, D, Bertero, A, Tavares, JM, Pedersen, RA, Vallier, L, et al.. Inducible and deterministic forward programming of human pluripotent stem cells into neurons, skeletal myocytes, and oligodendrocytes. Stem Cell Rep 2017;8:803–12. https://doi.org/10.1016/j.stemcr.2017.02.016.Search in Google Scholar PubMed PubMed Central
46. Moreau, T, Evans, AL, Vasquez, L, Tijssen, MR, Yan, Y, Trotter, MW, et al.. Large-scale production of megakaryocytes from human pluripotent stem cells by chemically defined forward programming. Nat Commun 2016;7:11208. https://doi.org/10.1038/ncomms11208.Search in Google Scholar PubMed PubMed Central
47. Dame, K, Cincotta, S, Lang, AH, Sanghrajka, RM, Zhang, L, Choi, J, et al.. Thyroid progenitors are robustly derived from embryonic stem cells through transient, developmental stage-specific overexpression of Nkx2-1. Stem Cell Rep 2017;8:216–25. https://doi.org/10.1016/j.stemcr.2016.12.024.Search in Google Scholar PubMed PubMed Central
48. Velazquez, JJ, LeGraw, R, Moghadam, F, Tan, Y, Kilbourne, J, Maggiore, JC, et al.. Gene regulatory network analysis and engineering directs development and vascularization of multilineage human liver organoids. Cell Syst 2021;12:41–55.e11. https://doi.org/10.1016/j.cels.2020.11.002.Search in Google Scholar PubMed PubMed Central
49. Wilson, HV. A new method by which sponges may be artificially reared. Science 1907;25:912–5. https://doi.org/10.1126/science.25.649.912.Search in Google Scholar PubMed
50. Sato, T, Vries, RG, Snippert, HJ, van de Wetering, M, Barker, N, Stange, DE, et al.. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262–5. https://doi.org/10.1038/nature07935.Search in Google Scholar PubMed
51. Miura, Y, Li, M-Y, Birey, F, Ikeda, K, Revah, O, Thete, MV, et al.. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat Biotechnol 2020;38:1421–30. https://doi.org/10.1038/s41587-020-00763-w.Search in Google Scholar PubMed PubMed Central
52. Lawlor, KT, Vanslambrouck, JM, Higgins, JW, Chambon, A, Bishard, K, Arndt, D, et al.. Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat Mater 2021;20:260–71. https://doi.org/10.1038/s41563-020-00853-9.Search in Google Scholar PubMed PubMed Central
53. Ouchi, R, Togo, S, Kimura, M, Shinozawa, T, Koido, M, Koike, H, et al.. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metabol 2019;30:374–84.e6. https://doi.org/10.1016/j.cmet.2019.05.007.Search in Google Scholar PubMed PubMed Central
54. Paşca, AM, Sloan, SA, Clarke, LE, Tian, Y, Makinson, CD, Huber, N, et al.. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 2015;12:671–8. https://doi.org/10.1038/nmeth.3415.Search in Google Scholar PubMed PubMed Central
55. Huang, WK, Wong, SZH, Pather, SR, Nguyen, PTT, Zhang, F, Zhang, DY, et al.. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell 2021;28:1657–70.e10. https://doi.org/10.1016/j.stem.2021.04.006.Search in Google Scholar PubMed PubMed Central
56. Garreta, E, Kamm, RD, Chuva de Sousa Lopes, SM, Lancaster, MA, Weiss, R, Trepat, X, et al.. Rethinking organoid technology through bioengineering. Nat Mater 2021;20:145–55. https://doi.org/10.1038/s41563-020-00804-4.Search in Google Scholar PubMed
57. Cho, AN, Jin, Y, An, Y, Kim, J, Choi, YS, Lee, JS, et al.. Microfluidic device with brain extracellular matrix promotes structural and functional maturation of human brain organoids. Nat Commun 2021;12:4730. https://doi.org/10.1038/s41467-021-24775-5.Search in Google Scholar PubMed PubMed Central
58. Kagawa, H, Javali, A, Khoei, HH, Sommer, TM, Sestini, G, Novatchkova, M, et al.. Human blastoids model blastocyst development and implantation. Nature 2022;601:600–5. https://doi.org/10.1038/s41586-021-04267-8.Search in Google Scholar PubMed PubMed Central
59. Baker, DJ, Arany, Z, Baur, JA, Epstein, JA, June, CH. CAR T therapy beyond cancer: the evolution of a living drug. Nature 2023;619:707–15. https://doi.org/10.1038/s41586-023-06243-w.Search in Google Scholar PubMed
60. Kroeze, WK, Sassano, MF, Huang, XP, Lansu, K, McCorvy, JD, Giguère, PM, et al.. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol 2015;22:362–9. https://doi.org/10.1038/nsmb.3014.Search in Google Scholar PubMed PubMed Central
61. Kipniss, NH, Dingal, P, Abbott, TR, Gao, Y, Wang, H, Dominguez, AA, et al.. Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system. Nat Commun 2017;8:2212. https://doi.org/10.1038/s41467-017-02075-1.Search in Google Scholar PubMed PubMed Central
62. Daringer, NM, Dudek, RM, Schwarz, KA, Leonard, JN. Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth Biol 2014;3:892–902. https://doi.org/10.1021/sb400128g.Search in Google Scholar PubMed PubMed Central
63. Morsut, L, Roybal, KT, Xiong, X, Gordley, RM, Coyle, SM, Thomson, M, et al.. Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell 2016;164:780–91. https://doi.org/10.1016/j.cell.2016.01.012.Search in Google Scholar PubMed PubMed Central
64. Zhu, I, Liu, R, Garcia, JM, Hyrenius-Wittsten, A, Piraner, DI, Alavi, J, et al.. Modular design of synthetic receptors for programmed gene regulation in cell therapies. Cell 2022;185:1431–43 e16. https://doi.org/10.1016/j.cell.2022.03.023.Search in Google Scholar PubMed PubMed Central
65. Saxena, P, Heng, BC, Bai, P, Folcher, M, Zulewski, H, Fussenegger, M. A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells. Nat Commun 2016;7:11247. https://doi.org/10.1038/ncomms11247.Search in Google Scholar PubMed PubMed Central
66. Miki, K, Endo, K, Takahashi, S, Funakoshi, S, Takei, I, Katayama, S, et al.. Efficient detection and purification of cell populations using synthetic MicroRNA switches. Cell Stem Cell 2015;16:699–711. https://doi.org/10.1016/j.stem.2015.04.005.Search in Google Scholar PubMed
67. Strati, P, Bachanova, V, Goodman, A, Pagel, JM, Castro, JE, Griffis, K, et al.. Preliminary results of a phase I trial of FT516, an off-the-shelf natural killer (NK) cell therapy derived from a clonal master induced pluripotent stem cell (iPSC) line expressing high-affinity, non-cleavable CD16 (hnCD16), in patients (pts) with relapsed/refractory (R/R) B-cell lymphoma (BCL). J Clin Oncol 2021;39:7541. https://doi.org/10.1200/jco.2021.39.15_suppl.7541.Search in Google Scholar
68. Woan, KV, Kim, H, Bjordahl, R, Davis, ZB, Gaidarova, S, Goulding, J, et al.. Harnessing features of adaptive NK cells to generate iPSC-derived NK cells for enhanced immunotherapy. Cell Stem Cell 2021;28:2062–75 e5. https://doi.org/10.1016/j.stem.2021.08.013.Search in Google Scholar PubMed PubMed Central
69. Goldenson, BH, Kaufman, DS. Into the multiverse of gene edited NK cell-based therapeutic strategies. Cell Stem Cell 2021;28:2041–3. https://doi.org/10.1016/j.stem.2021.11.004.Search in Google Scholar PubMed
70. Li, Y, Hermanson, DL, Moriarity, BS, Kaufman, DS. Human iPSC-derived natural killer cells engineered with chimeric antigen receptors enhance anti-tumor activity. Cell Stem Cell 2018;23:181–92 e5. https://doi.org/10.1016/j.stem.2018.06.002.Search in Google Scholar PubMed PubMed Central
71. Zhu, H, Blum, RH, Bernareggi, D, Ask, EH, Wu, Z, Hoel, HJ, et al.. Metabolic reprograming via deletion of CISH in human iPSC-derived NK cells promotes in vivo persistence and enhances anti-tumor activity. Cell Stem Cell 2020;27:224–37. https://doi.org/10.1016/j.stem.2020.05.008.Search in Google Scholar PubMed PubMed Central
72. Scheller, L, Strittmatter, T, Fuchs, D, Bojar, D, Fussenegger, M. Generalized extracellular molecule sensor platform for programming cellular behavior. Nat Chem Biol 2018;14:723–9. https://doi.org/10.1038/s41589-018-0046-z.Search in Google Scholar PubMed
73. Ji, TT, Niu, SS, Fang, MH, Xu, LX, Wang, X, Zou, J, et al.. Genome editing HLA alleles for a pilot immunocompatible hESC line in a Chinese hESC bank for cell therapies. Cell Prolif 2023;56:e13471. https://doi.org/10.1111/cpr.13471.Search in Google Scholar PubMed PubMed Central
74. Toda, S, Blauch, LR, Tang, SKY, Morsut, L, Lim, WA. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 2018;361:156–62. https://doi.org/10.1126/science.aat0271.Search in Google Scholar PubMed PubMed Central
75. Zhang, S, Zhao, H, Liu, Z, Liu, K, Zhu, H, Pu, W, et al.. Monitoring of cell-cell communication and contact history in mammals. Science 2022;378:eabo5503. https://doi.org/10.1126/science.abo5503.Search in Google Scholar PubMed
76. Malaguti, M, Portero Migueles, R, Annoh, J, Sadurska, D, Blin, G, Lowell, S. SyNPL: synthetic Notch pluripotent cell lines to monitor and manipulate cell interactions in vitro and in vivo. Development 2022;149:dev200226. https://doi.org/10.1242/dev.200226.Search in Google Scholar PubMed PubMed Central
77. Toda, S, McKeithan, WL, Hakkinen, TJ, Lopez, P, Klein, OD, Lim, WA. Engineering synthetic morphogen systems that can program multicellular patterning. Science 2020;370:327–31. https://doi.org/10.1126/science.abc0033.Search in Google Scholar PubMed PubMed Central
78. Repina, NA, Johnson, HJ, Bao, X, Zimmermann, JA, Joy, DA, Bi, SZ, et al.. Optogenetic control of Wnt signaling models cell-intrinsic embryogenic patterning using 2D human pluripotent stem cell culture. Development 2023;150:dev201386. https://doi.org/10.1242/dev.201386.Search in Google Scholar PubMed PubMed Central
79. Sako, K, Pradhan, SJ, Barone, V, Inglés-Prieto, Á, Müller, P, Ruprecht, V, et al.. Optogenetic control of nodal signaling reveals a temporal pattern of nodal signaling regulating cell fate specification during gastrulation. Cell Rep 2016;16:866–77. https://doi.org/10.1016/j.celrep.2016.06.036.Search in Google Scholar PubMed
80. Humphreys, PA, Woods, S, Smith, CA, Bates, N, Cain, SA, Lucas, R, et al.. Optogenetic control of the BMP signaling pathway. ACS Synth Biol 2020;9:3067–78. https://doi.org/10.1021/acssynbio.0c00315.Search in Google Scholar PubMed PubMed Central
81. Chao, G, Wannier, TM, Gutierrez, C, Borders, NC, Appleton, E, Chadha, A, et al.. helixCAM: a platform for programmable cellular assembly in bacteria and human cells. Cell 2022;185:3551–67.e39. https://doi.org/10.1016/j.cell.2022.08.012.Search in Google Scholar PubMed PubMed Central
82. Glass, DS, Riedel-Kruse, IH. A synthetic bacterial cell-cell adhesion toolbox for programming multicellular morphologies and patterns. Cell 2018;174:649–58.e16. https://doi.org/10.1016/j.cell.2018.06.041.Search in Google Scholar PubMed
83. Stevens, AJ, Harris, AR, Gerdts, J, Kim, KH, Trentesaux, C, Ramirez, JT, et al.. Programming multicellular assembly with synthetic cell adhesion molecules. Nature 2023;614:144–52. https://doi.org/10.1038/s41586-022-05622-z.Search in Google Scholar PubMed PubMed Central
84. Bao, M, Cornwall-Scoones, J, Sanchez-Vasquez, E, Cox, AL, Chen, DY, De Jonghe, J, et al.. Stem cell-derived synthetic embryos self-assemble by exploiting cadherin codes and cortical tension. Nat Cell Biol 2022;24:1341–9. https://doi.org/10.1038/s41556-022-00984-y.Search in Google Scholar PubMed PubMed Central
85. Joung, J, Ma, S, Tay, T, Geiger-Schuller, KR, Kirchgatterer, PC, Verdine, VK, et al.. A transcription factor atlas of directed differentiation. Cell 2023;186:209–29 e26. https://doi.org/10.1016/j.cell.2022.11.026.Search in Google Scholar PubMed PubMed Central
86. Rukhlenko, OS, Halasz, M, Rauch, N, Zhernovkov, V, Prince, T, Wynne, K, et al.. Control of cell state transitions. Nature 2022;609:975–85. https://doi.org/10.1038/s41586-022-05194-y.Search in Google Scholar PubMed PubMed Central
87. Bethel, NP, Borst, AJ, Parmeggiani, F, Bick, MJ, Brunette, TJ, Nguyen, H, et al.. Precisely patterned nanofibres made from extendable protein multiplexes. Nat Chem 2023;15:1664–71. https://doi.org/10.1038/s41557-023-01314-x.Search in Google Scholar PubMed PubMed Central
88. Xie, M, Ye, H, Wang, H, Charpin-El Hamri, G, Lormeau, C, Saxena, P, et al.. β-cell–mimetic designer cells provide closed-loop glycemic control. Science 2016;354:1296–301. https://doi.org/10.1126/science.aaf4006.Search in Google Scholar PubMed
89. Beitz, A, Oakes, C, Galloway, K. Synthetic gene circuits as tools for drug discovery. Trends Biotechnol 2022;40:210–25. https://doi.org/10.1016/j.tibtech.2021.06.007.Search in Google Scholar PubMed
90. Frei, T, Cella, F, Tedeschi, F, Gutiérrez, J, Stan, GB, Khammash, M, et al.. Characterization and mitigation of gene expression burden in mammalian cells. Nat Commun 2020;11:4641. https://doi.org/10.1038/s41467-020-18392-x.Search in Google Scholar PubMed PubMed Central
91. Zhou, Z, Liu, Y, Feng, Y, Klepin, S, Tsimring, LS, Pillus, L, et al.. Engineering longevity—design of a synthetic gene oscillator to slow cellular aging. Science 2023;380:376–81. https://doi.org/10.1126/science.add7631.Search in Google Scholar PubMed PubMed Central
92. Zhu, R, Del Rio-Salgado, JM, Garcia-Ojalvo, J, Elowitz, MB. Synthetic multistability in mammalian cells. Science 2022;375:eabg9765. https://doi.org/10.1126/science.abg9765.Search in Google Scholar PubMed
93. Han, J, Zhu, L, Zhang, J, Guo, L, Sun, X, Huang, C, et al.. Rational engineering of adeno-associated virus capsid enhances human hepatocyte tropism and reduces immunogenicity. Cell Prolif 2022;55:e13339. https://doi.org/10.1111/cpr.13339.Search in Google Scholar PubMed PubMed Central
94. Li, C, Samulski, RJ. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet 2020;21:255–72. https://doi.org/10.1038/s41576-019-0205-4.Search in Google Scholar PubMed
95. Bost, JP, Barriga, H, Holme, MN, Gallud, A, Maugeri, M, Gupta, D, et al.. Delivery of oligonucleotide therapeutics: chemical modifications, lipid nanoparticles, and extracellular vesicles. ACS Nano 2021;15:13993–4021. https://doi.org/10.1021/acsnano.1c05099.Search in Google Scholar PubMed PubMed Central
96. Ozelo, MC, Mahlangu, J, Pasi, KJ, Giermasz, A, Leavitt, AD, Laffan, M, et al.. Valoctocogene roxaparvovec gene therapy for hemophilia A. N Engl J Med 2022;386:1013–25. https://doi.org/10.1056/nejmoa2113708.Search in Google Scholar
97. Gillmore, JD, Gane, E, Taubel, J, Kao, J, Fontana, M, Maitland, ML, et al.. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N Engl J Med 2021;385:493–502. https://doi.org/10.1056/nejmoa2107454.Search in Google Scholar PubMed
98. Peng, YJ, Huang, X, Zhou, Q. Ethical and policy considerations for human embryo and stem cell research in China. Cell Stem Cell 2020;27:511–4. https://doi.org/10.1016/j.stem.2020.09.010.Search in Google Scholar PubMed
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorial
- Opportunity and challenge of rehabilitation medicine in China
- Reviews
- Engineering pluripotent stem cells with synthetic biology for regenerative medicine
- Assembling the RNA therapeutics toolbox
- Organoids as preclinical models of human disease: progress and applications
- Perspectives
- The role of bile acids in human aging
- Hormone-based pharmacotherapy for metabolic dysfunction-associated fatty liver disease
- Letter
- Clinical characteristics and pharmacokinetics of PAXLOVID in COVID-19 patients with hematological tumor
Articles in the same Issue
- Frontmatter
- Editorial
- Opportunity and challenge of rehabilitation medicine in China
- Reviews
- Engineering pluripotent stem cells with synthetic biology for regenerative medicine
- Assembling the RNA therapeutics toolbox
- Organoids as preclinical models of human disease: progress and applications
- Perspectives
- The role of bile acids in human aging
- Hormone-based pharmacotherapy for metabolic dysfunction-associated fatty liver disease
- Letter
- Clinical characteristics and pharmacokinetics of PAXLOVID in COVID-19 patients with hematological tumor


