Abstract
The organoid field has been developing rapidly during the last decade. Organoids for human pre-, peri- and post-implantation development have opened an avenue to study these biological processes in vitro, which have been hampered by lack of accessible research models for long term. The technologies of four fields, single cell omics sequencing, genome editing and lineage tracing, microfluidics and tissue engineering, have fueled the rapid development of the organoid field. In this review, we will discuss the organoid research on human early development as well as future directions of the organoid field combining with other powerful technologies.
Introduction of organoids
Cell self-organization is a fundamental nature of animal development in vivo. The development of a whole human body from a fertilized egg can be regarded as a highly complex and highly ordered self-organization process. Two-dimensional (2D) cell culturing has made great contributions to our knowledge of nearly all basic biological processes such as DNA replication, transcription, translation, cell cycling and signaling transduction. However, the 2D cultured cells usually show no self-organization characteristics as they do during development. An organoid is defined as a structure in which pluripotent stem cells (PSCs), multipotent stem cells, or adult stem cells (ASCs) are differentiated into multiple cell populations that self-organize or assemble into a tissue that resembles a mini-organ in vivo in three-dimensional (3D) culture. Yoshiki Sasai’s group and Hans Cleves’ group are the first to show that, when cultured in 3D conditions, pluripotent stem cells (PSCs) and adult stem cells (ASCs) are able to self-organize into mini-organ-like structures. Sasai’s group showed that aggregates of mouse embryonic stem cells (ESCs) in 3D culture were able to autonomously generate polarized cortical neuroepithelia, elegant optic cups and anterior pituitary structures [1], [2], [3]. At the same time, Sato et al. in Cleves’ group showed that a single Lgr5-positive murine intestine stem cell cultured in 3D matrigel was able to form an intestine crypt–villus structure [4]. These works demonstrated the remarkable self-organization capacity of in vitro cultured cells and opened the avenue of the organoid field. In the last decade, the organoid field has been boosting [5, 6]. Since the work of Sasai, Clevers, Sato and colleagues, organoids for most mouse and human organs including brain, intestine, stomach, liver, lung, kidney, blood vessel and heart have been reported [7], [8], [9], [10], [11], [12], [13]. In this review, we will focus on organoids of mammalian (particularly human) early development, which are also termed as embryoid, blastoid or gastruloid for different structures. We will also discuss on future directions of the organoid field in general.
Organoids of mammalian early development
The mammalian early development, including pre-, peri- and post-implantation development, starts from the fertilized egg or the zygote. After several rounds of cleavage divisions and two waves of asymmetric divisions, a zygote forms a blastocyst which then implants into the uterus. Then, a series of complex morphological changes including polarization and lumenogenesis of the epiblast, primitive streaking and gastrulation occur for building three germ layers of the body, as well as primordial germ cells which is generated to faithfully transmit genetic information from generation to generation to complete the life cycle of an individual [14]. Thus, the mammalian early development is a natural self-organization process. Since the nutritious and oxygen problem is still not serious during the stage due to the relatively small size of the embryos, one can imagine that it is possible to reconstitute parts or even whole of this process in vitro using cultured stem cells [15, 16].
The blastocyst is the first well-organized structure in mammalian early development. It contains three cell types: the outer extra-embryonic trophectoderm (TE), the inner pluripotent epiblast (EPI) and primitive endoderm (PE). In morphology, it contains a fluid-filled cavity within the spherical thin-walled TE cells. For reconstituting a blastocyst-like structure, Nicolas et al. reached the first success in 2018 by assembly of mouse ESCs and trophectoderm stem cells (TSCs) [17]. In the approach, mouse ESCs were firstly added to microwells to allow formation of aggregates and then mouse TSCs were added that leaded to spontaneous organization of TSC cysts with internal ESC aggregates, which was termed blastoids by the authors. Stimulation of cyclic adenosine monophosphate (cAMP) and Wingless/INT-1 (WNT) signaling pathway greatly increased TSC cavitation and blastoid formation, reaching efficiency as high as 70% at a certain combination ratio of 8 ESC cells and 20 TSC cells per microwell. Later, several groups generated mouse blastocyst-like structures using various protocols, including the one using extended pluripotent stem cells as the single starting cell type [18], [19], [20], [21]. In 2021, several groups reported the generation of human blastocyst-like structures [22], [23], [24], [25], [26], [27]. In one of these studies also by Nicolas and colleagues, naive human pluripotent stem cells (hPSCs) were aggregated in non-adherent hydrogel microwells upon inhibition of the Hippo, transforming growth Factor β (TGF-β) and extracellular regulated protein kinases (ERK) pathways [24]. They showed that this approach was able to generate human blastoids with more than 70% efficiency.
Implantation is a highly complex maternal-fetal interaction process including attachment of the blastocyst to the uterine surface epithelium, mature of polar TE, invasion of the trophoblast into endometrium, and transformation of endometrium into decidua [28]. The mouse and human blastoids show key characteristics for implantation. After transferred into the uterus of pseudo-pregnant mice, blastoids were able to induce deciduae formation [17]. Also, the human blastoids were able to attach to hormonally stimulated endometrial cells [24]. Interestingly, results showed that pseudo-blastoids without epiblast component cannot attach, indicating that EPI induces polar TE maturation for attachment [24]. Except for the blastoids, researchers also developed other organoid systems for modeling implantation. Trophoblasts derived from human placentas can be cultured in Matrigel-based 3D system to form trophoblast organoids, which can be kept for long term and showed complex structures resembling the organization of placental villi in vivo [29, 30]. Also, Park et al. developed an implantation-on-a-chip micro-engineered system to model the invasion of fetal extravillous trophoblasts into the maternal uterus [31].
During peri-implantation development, EPI polarizes and forms a pro-amniotic cavity, and a bipolar embryonic sac is generated with specification of primitive streak cells setting for gastrulation. Bedzhov et al. showed that TE and PE derived laminin provided a basal membrane niche for polarization and lumenogenesis of EPI in ex vivo cultured mouse peri-implantation embryos [32, 33]. This process can be mimicked in cultured ESCs. When ESCs are cultured in a 2D environment with laminin, they form an epithelium structure. While they are dissociated and then put in a 3D environment with extracellular matrix (ECM) proteins, they will self-assemble into a lumen structure with an epithelium [32, 34]. The 3D cultured ESCs can also be transformed into an amnion-like squamous cyst structure with activated bone morphogenetic protein- (BMP)/small mother against decapentaplegic (SMAD) signaling [35]. Further, using a microfluidic device, Zheng et al. generated an asymmetric epiblast-like cyst structure resembling the human embryonic sac before the onset of gastrulation at 7 to 12 days post-fertilization [36].
Gastrulation is a highly ordered and complex process to build the animal body plan, together with differentiation of three germ layers: the ectoderm, mesendoderm and endoderm. While the above-mentioned organoid models recapitulate key aspects of peri-implantation development, none of them reach the gastrulation stage. It has long been known that suspended ESCs aggregate into a structure termed embryoid body (EB) that can differentiate into various cell types of all three germ lays. However, it shows little ordered self-organization features. So EBs were usually regarded as a disorganized embryo-like structure. Warmflash et al. demonstrated that colony size and geometry were important factors for reproducibly spatially ordered differentiation. When confining the colony size using micropatterns, human ESC colonies can robustly differentiate into outer trophectoderm-like, intermediate mesendoderm-like and an inner ectoderm-like cells, in response to BMP4 [37]. Using the micropatterned culture system, human ESCs were induced into a human organizer that was able to induce and contribute to a secondary axis when grafted into chick embryos [38]. To promote an ESC-derived cell aggregate to initiate a gastrulation-like event, one should first break symmetry of the aggregate. Simunovic et al. showed that a defined uniform BMP can spontaneously break symmetry of 3D-cultured human ESCs via activating both the WNT signaling and its inhibitor Dickkopf (DKK) [39]. Alfonso Martinez Arias and colleagues showed that mouse and human ESC aggregates, when pretreated by Wnt agonist before aggregating, were able to break symmetry and further undergo gastrulation-like events and elongation along an anterior-posterior axis [40], [41], [42]. This 3D structure, which they termed gastruloids, showed patterned expression of neural, mesodermal and endodermal cell types, and when put into a low concentration of Matrigel, induced somatogenesis [43]. Together, these results demonstrate the remarkable capacity of self-organization in vitro cultured ESCs that can mimic a developmental process as complex as gastrulation.
Future directions of the development of organoid systems
The organoid technology is closely associated or interconnected with technologies of other fields. Advances of single cell omics sequencing, genome editing and lineage tracing, microfluidics and tissue engineering technologies not only have supported and promoted the development of the organoid field during the last decade, but will also further fuel its future progressions (Figure 1).
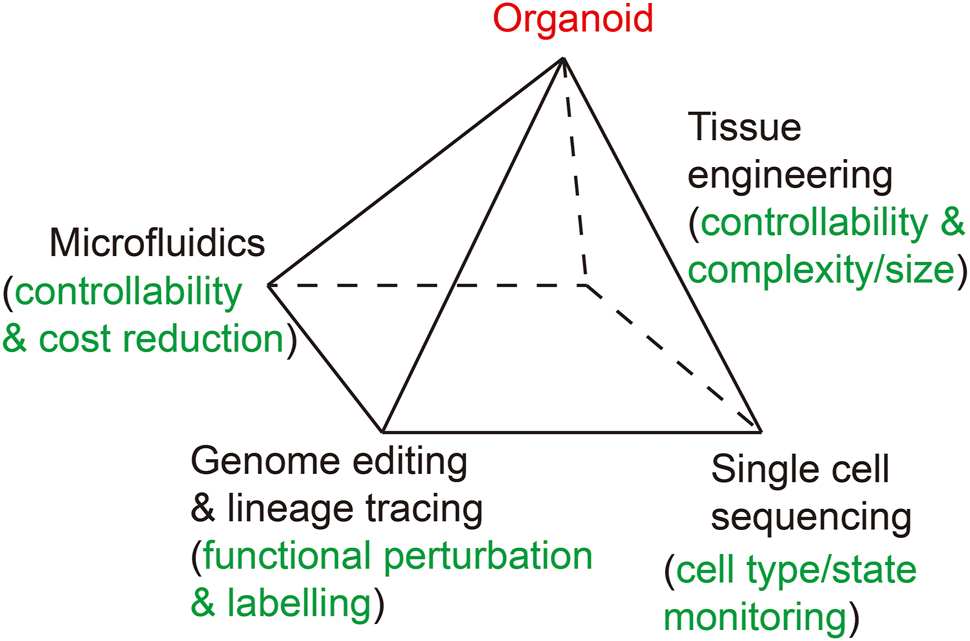
A diagram shows interconnection between the organoid field and other technologies. Four technologies including single cell omics sequencing, gene editing and lineage tracing, microfluidics and tissue engineering promote the organoid field.
Combining organoid systems with single cell omics sequencing technologies
For the organoid culture systems, the most valuable usage is to use them to mimic the in vivo embryonic development process under physiological conditions. Comparing with conventional 2D cell culture strategy, the cellular heterogeneities are greatly elevated in 3D organoid culture systems. Immunostaining is generally applied for organoids, always providing a first clue on whether a certain cell type exists in an organoid. However, even when a cell in culture expresses a number of marker genes specific to a certain cell type in vivo, one cannot make a conclusion that the given cell type is captured in the organoid. Furthermore, it is difficult to determine if all types of the cells in an organoid faithfully resemble those in an embryo of the exact developmental stage, the exact cell types, and the exact proportions and spatial organizations. In contrast to immunostaining, single cell RNA-seq provides a much higher dimensional view on the transcriptome state of individual cells, and thus information on how many cell types or cell states exist in the organoids and how closely the examined cell type in organoids resemble the corresponding cell type in vivo [44]. The Human Developmental Cell Atlas, which is a critical part of the Human Cell Atlas project, plans to give an accurate atlas of human embryonic development at single-cell resolution and whole-transcriptome scale [45]. Several scRNA-seq databases of the human and monkey pre-, peri-, and post-implantation development have been available during recently years, which provide invaluable references of comparison for evaluating the in vitro human early developmental organoid models [46], [47], [48], [49], [50], [51], [52], [53]. Niu et al. group also generated organoids from monkey blastocysts, which have great values for stem cell line derived organoid studies [52]. With these powerful resources in hand, we will know whether all cell types and their proportions and spatial organization in an organoid faithfully resemble those in an embryo. In this way, we will have a gold standard for the quality of the organoids from mimicking the in vivo embryonic development point of view. This will resolve many potential debates in the field, for example, among many different protocols and strategies from different labs to generate the same type of organoids, which is a better one. The one most faithfully mimic the in vivo developmental process based on single cell omics sequencing results will probably be the best one.
For human blastoids, scRNA-seq showed that they comprised only three distinct transcriptomic states, and these states matched with the TE, EPI and PE cell types in vivo, thus confirming their resembling to human blastocysts [24]. Also, scRNA-seq analysis of mouse gastruloids showed that the gastruloids contained various cell types from three germ layers; yet, the anterior neuronal cell types were absent [43]. For the ASC-derived organoids, as an example, scRNA-seq for mouse intestinal organoids distinguished cell groups corresponding to enterocytes, enterocyte precursors and transit amplifying cells, as well as rare enteroendocrine cells, thus confirming the differentiation features of the intestinal organoids [54]. These scRNA-seq data indicate that the organoids both closely resemble and have discernible differences with the corresponding type of cells in vivo.
Another layer of single cell omics is the epigenome. It comprises a complex set of features on genome that regulate gene expression in a DNA sequence-independent manner, including the chromatin accessibility, DNA methylation, histone modifications, transcription factor binding to genomic loci and chromatin structure conformation. Single cell epigenome sequencing techniques have been developed fast during recent years [55]. Epigenome should provide regulatory layers of information for comparisons between the organoid and the corresponding in vivo development. For example, the trophoblast organoids were shown to display global DNA hypomethylation and local hypermethylation patterns similar to in vivo TE, thus verifying their characteristics [29]. Since the transcriptome differences between the organoid cell types and their corresponding in vivo cell types usually have an epigenetic reason, the single cell epigenome sequencing should help elucidate this and thus provide clues for improving the organoid culture conditions. Recently single-cell chromatin accessibility data have identified approximately 1.2 million candidate cis-regulatory elements (cCREs) in 222 distinct cell types in human fetal and adult tissues, which enrich hundreds of genome wide association study (GWAS) variants of traits/diseases [56]. Combined analysis of human trait/disease modeling organoids and single-cell epigenome sequencing should help explore molecular mechanisms about how these GWAS variants are involved in human traits/diseases.
Further, the spatial transcriptomics technologies will help elucidate the spatially resolved gene expression patterns of an organoid structure [57]. A spatial transcriptomics technique, tomo-seq, has been applied to analyzing the mouse gastruloids. The results showed a reproducible expression pattern shift from the anterior to the posterior sections of the gastruloids, which was similar to the E8.5 mouse embryos in vivo, thus providing reliable evidence that the gastruloid showed anterior-to-posterior axis formation resembling the in vivo gastrulation development [43]. The organoid formation generally has much lower reproducibility comparing with their corresponding in vivo organs. This makes it more difficulty for the spatial transcriptomics analyses of an organoid comparing with a normal embryo or organ. The spatial transcriptomics technologies are rapidly developing regarding easiness, cost, throughput and particularly single-cell resolution. These improvements should increase their powerfulness for analysis of organoids.
Combining organoid systems with genome editing and lineage tracing technologies
The CRISPR/Cas-based genome editing technology has revolutionized the biological and medical fields. It provides very powerful tools for functional analysis to reveal the causal genotype-phenotype relationships at whole genome scale in one experiment, permitting thousands or even all of the twenty thousand protein-coding genes in our genome being functionally screened [58]. One of the most important advantages of the organoid is that a large number of subjects are available for functional screening, which alleviates the ethical problems related to acquiring human embryo or fetal tissues for experimental analyses. The combination of organoid systems and genome editing technologies will be an ideal strategy to address the functions of thousands of genes involved in human embryonic and fetal development.
However, one disadvantage of the organoid is the relatively high variabilities and high batch effects. To overcome these limitations, Esk et al. developed a strategy that compared the wild-type and the mutant cells within the same individual organoid by mixing the wild-type stem cells and the mutant stem cells before generating individual organoids, and screened for microcephaly candidate genes using cerebral organoids [59]. They also incorporated another powerful strategy, genetic lineage tracing, into the organoid analysis by tagging different barcodes to the wild-type and mutant (CRISPR)/Cas constructs. By counting the ratio of the barcodes of the mutant to the wild-type Cas-transfected cells, the authors were able to discern whether a candidate gene specifically affected cell proliferation after neurogenesis or cell proliferation when in 2D stem cell culture.
There are over one million cis-regulatory elements in human genome, including promoters, enhancers, insulators, and silencers. The cis-regulatory elements are the essential nodes of gene regulatory network that controls development, yet the function and mechanism of most cis-regulatory elements in human genome for embryonic development are still unexplored. The organoid is an excellent model for studying the function and mechanism of a cis-regulatory element during human development. However, since a cis-regulatory element usually play fine-tuned roles in gene expression regulation, the high variability of the organoid system impedes such analysis. Besides the strategy to compare the wild-type and the mutant cells in the same individual organoids, another strategy is to compare gene expression between the mutant allele and the wild-type allele (which serves as the most stringent control currently) in the same individual cell, since generally the cis-regulatory element solely control the expression of the gene on the same chromosome [60] (Figure 2). Although heterozygous single nucleotide polymorphism (SNP) information in the exons of the analyzed gene is needed, this is the most stringent way to remove experimental variations and batch effects for gene functional studies in organoid systems.
![Figure 2:
Allele-specific analysis of a cis-regulatory element alleviates high variation of the organoid system. For one gene locus within an individual cell, there are two alleles (allele A and B marked by single nucleotide polymorphism [SNP] C and T, respectively). Upper panel: The gene on each allele is activated by a distant enhancer (green box) under normal condition. Lower panel: When the enhancer on allele A is disrupted by gene editing (grey box with a cross), the gene on allele A is inactivated while the one on allele B keeps expression, which can be discerned by specific reduction of the transcript with SNP C. In this way, one can perform functional study for the relationship between a putative cis-regulatory element and a gene in a well-controlled manner at single-cell level despite of the high variation of the organoid system.](/document/doi/10.1515/mr-2022-0028/asset/graphic/j_mr-2022-0028_fig_002.jpg)
Allele-specific analysis of a cis-regulatory element alleviates high variation of the organoid system. For one gene locus within an individual cell, there are two alleles (allele A and B marked by single nucleotide polymorphism [SNP] C and T, respectively). Upper panel: The gene on each allele is activated by a distant enhancer (green box) under normal condition. Lower panel: When the enhancer on allele A is disrupted by gene editing (grey box with a cross), the gene on allele A is inactivated while the one on allele B keeps expression, which can be discerned by specific reduction of the transcript with SNP C. In this way, one can perform functional study for the relationship between a putative cis-regulatory element and a gene in a well-controlled manner at single-cell level despite of the high variation of the organoid system.
An interesting strategy is to study the evolutionary molecular mechanisms of the cis-regulatory elements using allele-specific expression in organoids. Human has many species-specific characters even comparing to non-human primates. The most striking one is the evolution of our brain. The evolutionary molecular mechanisms underlying these characters should on a large part be genetic changes of the cis-regulatory elements of brain development related genes. However, direct comparative analyses of the brain development in human and non-human primate is very limited. Agoglia et al. fused the induced pluripotent stem cells (iPSCs) of human and chimpanzee to generate a set of tetraploid hybrid stem cells, and successfully generate brain organoids from these cells [61]. By comparing the allele-specific gene expression and allele-specific epigenetic modifications, this platform was great to investigate the differences of cis-regulatory elements between human and chimpanzee, since the alleles of human and chimpanzee existed in the same individual cell and thus essentially all of the systematical experimental variations were eliminated.
Lineage tracing is a powerful approach for understanding the developmental relationships among different cell types. This is generally achieved in animal models based on genetic tools. Endogenous genomic changes information has recently opened the door of lineage tracing for human development [62, 63]. However, this requires enough somatic mutation information to be covered, and is suitable for human clinical samples but cannot be combined with functional studies. The organoid system provides a unique opportunity for the lineage tracing study for human development and related diseases. Genetic lineage tracing techniques based on transgenic fluorescent proteins, Cre-mediated recombination or CRISPR/Cas9-mediated genome editing have already been established for the animal model study [64]. Using tamoxifen-inducible Cre-mediated fluorescent protein knock-in alleles of LGR5 and KRT20, the LGR5-positive and KRT20-positive cells of colorectal cancer organoids were traced after xenotransplanted under the renal capsules of immune-deficient mice. This revealed that the self-renewal and differentiation of the LGR5-positive cells as potential cancer stem cells, general post-mitotic feature of the KRT20-positive cells and the plasticity of KRT20-positive cells to revert back to proliferative stem cells [65]. Further multiphoton microscopy-based live cell lineage tracing of individual LGR5-positive cells in subcutaneous xenograft of colorectal cancer organoids identified p27 as a marker for dormant LGR5-positive cells [66]. Besides these traditional genetic lineage tracing approaches, the recently developed CRISPR/Cas9-based genetic lineage tracing techniques are able to build a high-resolution map for viewing all branching of the developmental lineage tree by sequential introducing DNA barcodes to multiple endogenous or transposon-integrated loci [67, 68]. When the DNA barcodes can be transcribed, scRNA-seq is able to be integrated to provide reliable lineage tracing and developmental trajectory information [68]. These techniques are particularly useful for the PSC-derived organoids that have more complexity of many different cell types and states.
Combining organoid systems with microfluidics technologies
The current organoid culture systems generally require milliliter volumes of medium with expensive cytokines and growth factors and are expensive. Combination of organoid systems with microfluidics technologies will be an ideal strategy to reduce the cost for several orders of magnitude by culturing individual organoids in microliter volumes. More importantly, microfluidics systems will allow more accurate control of the culture condition by providing better organization of different compartments, better control of the concentration and duration of cytokine treatment, better control of shearing forces, etc. In this way, the variations among individual organoids can be minimized, allowing for better removing of batch effects.
Park et al. built a microfluidic device to model 3D structural organization of human maternal-fetal interfaces [31]. The device contains three parallel channels. The center channel was injected with ECM hydrogel to create a 3D hydrogel scaffold that mimics the maternal endometrium. One side channel was seeded with primary human uterine endothelial cells (ECs) that mimic a maternal spiral artery. The other side channel was seeded with human extravillous trophoblasts (EVTs) to mimic the tip of the invading trophoblasts. The invasion of EVTs and the remodeling of ECs can be clearly visualized in this platform, thus greatly facilitated investigation on biological questions such as how ECs, decidualized stromal cells and uterine natural killer (uNK) cells regulated EVT invasion, and how invading EVTs regulated spiral artery remodeling. Particularly, many aspects of these questions are human specific that are difficult to be studied in animal models.
Zheng et al. built a microfluidic device to model the post-implantation human embryo [36]. The device also contained three parallel channels, including a cell-loading channel and a chemical-induction channel that were separated by a center channel loaded with basement membrane matrix. This platform thus allowed for precise control of signaling factors that functioned on the cells, from the aspects of ligand-receptor direction, type of interaction, concentration, and others. On the other hand, the cells can also be controlled and genetically manipulated. Besides highly controllable, the experimental system was also reproducible, scalable and more cost effective. The authors showed that BMP4 from the induction channel was able to highly reproducibly generate flattened, amniotic ectoderm-like cells at the pole exposed to BMP4, and a stratified, epiblast-like epithelium at the other pole. This structure resembled the human post-implantation asymmetry embryonic sac.
Combining organoid systems with tissue engineering technologies
The current organoid system is still not satisfactory regarding the size, lifespan and complexity. Improving of these aspects should increase the application scenarios of the organoid system to address more biological questions and fit the clinical therapy requirement. Combing with tissue engineering strategies should help improvement on these aspects of organoid system.
Tissue engineering strategies have been used to increase the size and improve the efficiency of organoid culture. Due to lack of vasculature, most current organoid models are limited to a size of less than one millimeter (mm) in diameter. The spinning bioreactor can enhance diffusion of oxygen and nutrients, and has been applied to support the long-term growth of suspended brain organoids up to 4 mm in diameter [8]. To overcome the shortcoming of low throughput, bulky size and high cost of commercially available spinning bioreactors, an in-house made miniaturized multiwell spinning bioreactor from 3D printing has been reported [69]. Further enhancing oxygen delivery with precise control of O2 and CO2 levels and atmospheric gas pressure in roller culture has been shown to allow ex utero development of mouse embryos to the hindlimb formation stage (E11) [70].
Micropatterning to confine the cell number is a simple and efficient tissue engineering strategy to improve the reproducibility of organoids [17, 37, 71]. Also, in order to guide 3D differentiation of the embryoid body to the brain organoid in a suspending culture system, a poly(lactide-co-glycolide) copolymer (PLGA) fiber microfilaments have been used as a floating scaffold, which improves neuroectoderm formation and cortical development of the microfilament-engineered cerebral organoids [72].
Assembly of different cell types or organoid types has been used to make a combined organoid with increased complexities. The gradient of sonic hedgehog (SHH) signaling activity is critical for forebrain regionalization. To add this signal to the brain organoid, an hPSC line with an inducible SHH allele were firstly aggregated before adding wild-type hPSCs for aggregation into the embryoid body. After differentiation of the embryoid body to neuroectoderm, the SHH was activated to induce forebrain organoid with an ordered self-organized dorso-ventral and anterior-posterior positional axes [73]. In another study, two types of human neural spheroids resembling the dorsal or the ventral forebrain were assembled to model the migration of GABAergic interneurons from the ventral to dorsal forebrain with their functional integration into the dorsal forebrain microcircuits [74]. Also, human PSC-derived neural crest cells and human developing intestine organoids were combined to generate intestine organoids with incorporated enteric nervous system [75]. A kidney metanephron organoid with contiguously forming nephron structures were reconstructed by assembling PSC-derived nephron progenitors, PSC-derived ureteric buds and stromal progenitor population sorted from embryonic mouse kidneys [76].
3D bioprinting is a powerful tissue engineering tool to precisely control cell deposition and biocompatible materials [77]. Researchers have investigated the application of 3D bioprinting for building complex organoids [78, 79]. In one study, human PSCs were 3D-printed to a heart structure with two chambers and a vessel inlet, and a beating mini-heart can be generated after differentiation of the PSCs [80]. In another study, intestine ASCs were used as building blocks to be directly 3D-printed into extracellular matrices to generate gastrointestine-like structure with self-organized features such as lumens and tubular intestinal epithelia [81]. These studies show that centimeter-scale organoids can be produced by using 3D bioprinting. Organ structures of even larger size are possible to be constructed using improved bioink technologies or decellularized organ scaffolds [82, 83].
Applications of organoid systems
The organoid should firstly help more thorough understanding of basic developmental biology, particularly for human development. For example, the human implantation defects are a major cause of early pregnancy loss. However, due to lack of available research models, molecular mechanisms of this process remain largely elusive [84]. Systems such as blastoids provide invaluable tools for elucidate this process. Also, human brain is one of the most marvelous products of evolution. Due to its uniqueness, many features of human brain are not suitable or not able to be studied by animal models such as mouse. Human brain organoids provide excellent novel tools to address many biological questions about human brain development.
The organoid system also serves as an excellent tool for human disease modeling [8, 85, 86]. Many human diseases do not have suitable animal models, which not only hamper understanding of their molecular mechanisms, but also hamper drug development. Chromosome aneuploidy is a big problem for human reproductive health [87]. It occurs in human preimplantation embryos at a frequency much higher than in the mouse. A model for studying chromosome aneuploidy in human preimplantation embryos is lacking for many years. A human chromosome aneuploidy model was developed using micropatterned human organoids [88]. The results suggested that aneuploid cells were greatly reduced from embryonic germ layers, but not from the extraembryonic tissue, thus supporting the emerging notion that aneuploid mosaic embryos were able to give healthy baby due to selective depletion of aneuploidy cells in EPI.
Many neuropsychiatric diseases such as autism spectrum disorder (ASD) are elusive in pathogenesis, despite that a number of disease-associated genes have been identified by GWAS studies. These diseases lack appropriate animal models and human cerebral organoids are emerging as a promising tool for understanding them. In particularly, patient-derived iPSC-cerebral organoids have been established to help reveal common and various pathological mechanisms of these diseases [89, 90].
For drug development, the United States Food and Drug Administration (FDA) has recently approved the first drug for chronic acquired demyelinating polyneuropathy (CIDP) and multifocal motor neuropathy (MMN) to enter clinical trials based on its efficiency data on organoid models without preclinical data from big animal models. As for precision medicine, the system can also be used to understand underlying mechanisms behind drug response variations among different individuals [91].
The organoid system can also be used for modeling human cancers. The patient cancer tissue-derived organoid is emerging as a promising tool to test the chemotherapy or target therapy drugs for precision therapy of cancer [92]. We recently showed that organoids from human colorectal cancer tissues faithfully mimic gene expression signatures of cancer epithelial cells. Organoids from adjacent normal tissues showed loss of normal colon epithelial cell’s gene expression signatures, but acquired some cancer epithelial cell’s gene expression features. This indicates that the colorectal cancer organoid can be a reliable system for screening for anti-cancer drugs for colorectal cancer patients. However, the organoid from the adjacent normal tissue may not be suitable for pairwise screening of drugs specifically killing colorectal cancer cells but not normal colorectal epithelial cells of the same patient [93]. Organoids can not only model the biological characters of the cancer cells, but also interactions between the cancer cells and their microenvironment cells including fibroblasts and immune cells such as T cells, NK cells, and microphages [94]. In addition, cancer initiation can also be modeled using PSC-derived organoids [95].
The organoid should also be applied to regeneration medicine. For example, current protocols for generating mature functional cells such as human pancreatic beta cells are mainly based on 2D culture. It is tempting to speculate that 3D organoid culture system will help establish more efficient protocol and more mature cells that are functionally closer to corresponding cells in vivo.
Perspectives
In the future, the organoid field should be improved in several aspects (Figure 3).

Characteristics of the current organoid technology. A pie chart shows the current states of seven major technical characters of current organoid systems in general: Reproducibility, controllability, throughput, cost, complexity, lifespan and size. The character is positioned more peripherally if it is in a more developed state.
First, improving the reproducibility. A great advantage of the animal model is that the in vivo biological processes, particularly development, are highly robust and reproducible. The robustness and reproducibility of current organoid systems are much lower and far from satisfying. The variations between different batches of organoid differentiation experiments are usually quite high even in the same lab from the same operator. And even the variations (morphologies, sizes, organizations, differentiation stages, etc.) among different organoids in the same dish are relatively high. So now mainly the master regulators for a specific developmental process are tested in organoid systems, leaving subtler phenotypes but probably more pathologically relevant regulators untested. The development is a well-controlled multiple-step process that any error at any step may be amplified, leading to variations between different individuals. The precise control of every step of organoid experiments is important, including the initial cell input, the number of the cells used, the timing and the concentration of signaling factors used, etc.
Second, improving the controllability. The in vivo tissue is composed of multiple cell types. These cell types interact with each other by reciprocal signaling pathways and extracellular matrix that have strict spatial distributions and fine-tuned functional organizations. It is a challenge to precisely control these inter-cellular interactions and spatial signaling in the organoid system, considering that we know very little about the self-assembling principles of embryonic development. Tissue engineering techniques such as microfluidics have been used for better controllability. Commercialization may help popularize these technologies.
Third, improving the throughput. One advantage of the organoid system is availability of large number of organoids that facilitate large scale functional screening. Further development of technologies for parallel analysis of the perturbation of hundreds or even thousands of genes should further enhance the power of organoid system for studying gene function under physiological conditions.
Fourth, reducing the cost. The cost of organoid culture is still high mainly due to using the expensive purified growth factors and cytokines as well as milliliter volume of the culture medium. Using small chemical molecules to replace the growth factor proteins may drastically reduce the cost. Also current ECM materials are generally made from Ewbing sarcomas with ECM proteins and growth factors. Is it possible to replace them with cheaper matrix. Microchips can help reduce the cost by reducing the reaction volume of the system from milliliters in a dish to microliters or even nanoliters in microfluidics chips.
Fifth, increasing the tissue complexity. PSC-derived organoids can contain cell types of all three germ layers; however, they are generally less complex and much less ordered than the corresponding tissues in vivo. The vasculature is not incorporated, which still serves as a big challenge in the field. Interactions with immune cells are missing, and stem cell niches are uncompleted. The left-right axis has not been mimicked as the anterior-posterior and dorsal-ventral axes. By assembling different cell types, tissues or organs and by applying tissue engineering tools including micropatterns, bioreactors and 3D bioprinting, we should be able to resemble more complex developmental processes and physiological functions in the organoid system, and build more complex and functional tissue and organ structures.
Sixth, increasing the lifespan. Comparing with animal models such as mouse, human organs have much longer period of lifespan including maturation and maintenance. For fully resembling physiological processes such as neuronal circuit construction and pathological processes such as senility, the organoid should have long enough lifespan. On the other hand, the molecular mechanisms underlying the different developmental duration of different species are elusive, and the organoid serves as a good model for investigation.
Seventh, increasing the size. Lacking vessel supports is a main limit factor of the sizes of the individual organoids. By using spinning to provide nutrient and oxygen, brain organoids can grow up to 4 mm of diameter. However, this is usually still much smaller than the in vivo organs. Improvement of culture system to allow more efficient nutrient and oxygen supply or even vessel development may enable development of larger organoids for more complex organ-level study and regeneration medicine. Ideally the size of an organoid should be improved from the current sub-millimeter to centimeter, which will mimic the development of an organ in a much better way. The achievement in tissue engineering field will help a lot for improving the complexity and sizes of organoids, which will probably permit reconstructing a functional human organ such as heart, lung, liver and kidney in the future [82].
Now organoid systems are helping us to mimic human embryonic development at millimeter scale, which permit us to analyze molecular mechanisms of numerous local interactions and self-organizations among different types of cells and at different developmental time points. In long-term, the organoid systems will probably be improved to mimic human embryonic and fetal development at centimeter or even whole-organ scale. Then it will be merged with tissue engineering technologies and will potentially permit us to reconstruct a whole organ with physiological functions. When we can de novo reconstruct an intact organ such as a liver, kidney, heart, or lung with physiological functions, or even a whole fetus, then we will know all of the main principles of our developmental biology. Just as Richard Feynman said: ‘What I cannot create, I do not understand’. When we can create an intact organ with physiological functions, or an intact fetus in vitro, we will really understand the core principles of how it is generated and how it works. Probably within next two or three decades, we will achieve it.
-
Research funding: We appreciate the support from the National Key R&D Program of China (2018YFA0107601).
-
Author contributions: All authors have accepted respon sibility for the entire content of this manuscript and approved its submission.
-
Competing interests: We declare no competing interests.
References
We apologize to the authors whose related great papers cannot be cited here due the space limitation.
1. Eiraku, M, Watanabe, K, Matsuo-Takasaki, M, Kawada, M, Yonemura, S, Matsumura, M, et al.. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008;3:519–32. https://doi.org/10.1016/j.stem.2008.09.002.Search in Google Scholar PubMed
2. Eiraku, M, Takata, N, Ishibashi, H, Kawada, M, Sakakura, E, Okuda, S, et al.. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011;472:51–6. https://doi.org/10.1038/nature09941.Search in Google Scholar PubMed
3. Suga, H, Kadoshima, T, Minaguchi, M, Ohgushi, M, Soen, M, Nakano, T, et al.. Self-formation of functional adenohypophysis in three-dimensional culture. Nature 2011;480:57–62. https://doi.org/10.1038/nature10637.Search in Google Scholar PubMed
4. Sato, T, Vries, RG, Snippert, HJ, van de Wetering, M, Barker, N, Stange, DE, et al.. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262–5. https://doi.org/10.1038/nature07935.Search in Google Scholar PubMed
5. Schutgens, F, Clevers, H. Human organoids: tools for understanding biology and treating diseases. Annu Rev Pathol 2020;15:211–34. https://doi.org/10.1146/annurev-pathmechdis-012419-032611.Search in Google Scholar PubMed
6. Corsini, NS, Knoblich, JA. Human organoids: new strategies and methods for analyzing human development and disease. Cell 2022;185:2756–69. https://doi.org/10.1016/j.cell.2022.06.051.Search in Google Scholar PubMed
7. Spence, JR, Mayhew, CN, Rankin, SA, Kuhar, MF, Vallance, JE, Tolle, K, et al.. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011;470:105–9. https://doi.org/10.1038/nature09691.Search in Google Scholar PubMed PubMed Central
8. Lancaster, MA, Renner, M, Martin, CA, Wenzel, D, Bicknell, LS, Hurles, ME, et al.. Cerebral organoids model human brain development and microcephaly. Nature 2013;501:373–9. https://doi.org/10.1038/nature12517.Search in Google Scholar PubMed PubMed Central
9. McCracken, KW, Catá, EM, Crawford, CM, Sinagoga, KL, Schumacher, M, Rockich, BE, et al.. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014;516:400–4. https://doi.org/10.1038/nature13863.Search in Google Scholar PubMed PubMed Central
10. Takebe, T, Sekine, K, Enomura, M, Koike, H, Kimura, M, Ogaeri, T, et al.. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013;499:481–4. https://doi.org/10.1038/nature12271.Search in Google Scholar PubMed
11. Chen, YW, Huang, SX, de Carvalho, A, Ho, SH, Islam, MN, Volpi, S, et al.. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 2017;19:542–9. https://doi.org/10.1038/ncb3510.Search in Google Scholar PubMed PubMed Central
12. Wimmer, RA, Leopoldi, A, Aichinger, M, Wick, N, Hantusch, B, Novatchkova, M, et al.. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 2019;565:505–10. https://doi.org/10.1038/s41586-018-0858-8.Search in Google Scholar PubMed PubMed Central
13. Hofbauer, P, Jahnel, SM, Papai, N, Giesshammer, M, Deyett, A, Schmidt, C, et al.. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 2021;184:3299–317.e22. https://doi.org/10.1016/j.cell.2021.04.034.Search in Google Scholar PubMed
14. Wen, L, Tang, F. Human germline cell development: from the perspective of single-cell sequencing. Mol Cell 2019;76:320–8. https://doi.org/10.1016/j.molcel.2019.08.025.Search in Google Scholar PubMed
15. Tarazi, S, Aguilera-Castrejon, A, Joubran, C, Ghanem, N, Ashouokhi, S, Roncato, F, et al.. Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs. Cell 2022;185:3290–306.e25. https://doi.org/10.1016/j.cell.2022.07.028.Search in Google Scholar PubMed PubMed Central
16. Amadei, G, Handford, CE, Qiu, C, De Jonghe, J, Greenfeld, H, Tran, M, et al.. Synthetic embryos complete gastrulation to neurulation and organogenesis. Nature 2022;610:143–53. https://doi.org/10.1038/s41586-022-05246-3.Search in Google Scholar PubMed PubMed Central
17. Rivron, NC, Frias-Aldeguer, J, Vrij, EJ, Boisset, JC, Korving, J, Vivié, J, et al.. Blastocyst-like structures generated solely from stem cells. Nature 2018;557:106–11. https://doi.org/10.1038/s41586-018-0051-0.Search in Google Scholar PubMed
18. Li, R, Zhong, C, Yu, Y, Liu, H, Sakurai, M, Yu, L, et al.. Generation of blastocyst-like structures from mouse embryonic and adult cell cultures. Cell 2019;179:687–702.e18. https://doi.org/10.1016/j.cell.2019.09.029.Search in Google Scholar PubMed PubMed Central
19. Kime, C, Kiyonari, H, Ohtsuka, S, Kohbayashi, E, Asahi, M, Yamanaka, S, et al.. Induced 2C expression and implantation-competent blastocyst-like cysts from primed pluripotent stem cells. Stem Cell Rep 2019;13:485–98. https://doi.org/10.1016/j.stemcr.2019.07.011.Search in Google Scholar PubMed PubMed Central
20. Sozen, B, Cox, AL, De Jonghe, J, Bao, M, Hollfelder, F, Glover, DM, et al.. Self-organization of mouse stem cells into an extended potential blastoid. Dev Cell 2019;51:698–712.e8. https://doi.org/10.1016/j.devcel.2019.11.014.Search in Google Scholar PubMed
21. Liu, K, Xu, X, Bai, D, Li, Y, Zhang, Y, Jia, Y, et al.. Bilineage embryo-like structure from EPS cells can produce live mice with tetraploid trophectoderm. Protein Cell 2022, pwac029. https://doi.org/10.1093/procel/pwac029 [Epub ahead of print].Search in Google Scholar
22. Yu, L, Wei, Y, Duan, J, Schmitz, DA, Sakurai, M, Wang, L, et al.. Blastocyst-like structures generated from human pluripotent stem cells. Nature 2021;591:620–6. https://doi.org/10.1038/s41586-021-03356-y.Search in Google Scholar PubMed
23. Liu, X, Tan, JP, Schröder, J, Aberkane, A, Ouyang, JF, Mohenska, M, et al.. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 2021;591:627–32. https://doi.org/10.1038/s41586-021-03372-y.Search in Google Scholar PubMed
24. Kagawa, H, Javali, A, Khoei, HH, Sommer, TM, Sestini, G, Novatchkova, M, et al.. Human blastoids model blastocyst development and implantation. Nature 2022;601:600–5. https://doi.org/10.1038/s41586-021-04267-8.Search in Google Scholar PubMed PubMed Central
25. Yanagida, A, Spindlow, D, Nichols, J, Dattani, A, Smith, A, Guo, G. Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell 2021;28:1016–22.e4. https://doi.org/10.1016/j.stem.2021.04.031.Search in Google Scholar PubMed PubMed Central
26. Sozen, B, Jorgensen, V, Weatherbee, BAT, Chen, S, Zhu, M, Zernicka-Goetz, M. Reconstructing aspects of human embryogenesis with pluripotent stem cells. Nat Commun 2021;12:5550. https://doi.org/10.1038/s41467-021-25853-4.Search in Google Scholar PubMed PubMed Central
27. Fan, Y, Min, Z, Alsolami, S, Ma, Z, Zhang, E, Chen, W, et al.. Generation of human blastocyst-like structures from pluripotent stem cells. Cell Discov 2021;7:81. https://doi.org/10.1038/s41421-021-00316-8.Search in Google Scholar PubMed PubMed Central
28. Turco, MY, Moffett, A. Development of the human placenta. Development 2019;146:dev163428. https://doi.org/10.1242/dev.163428.Search in Google Scholar PubMed
29. Turco, MY, Gardner, L, Kay, RG, Hamilton, RS, Prater, M, Hollinshead, MS, et al.. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 2018;564:263–7. https://doi.org/10.1038/s41586-018-0753-3.Search in Google Scholar PubMed PubMed Central
30. Haider, S, Meinhardt, G, Saleh, L, Kunihs, V, Gamperl, M, Kaindl, U, et al.. Self-renewing trophoblast organoids recapitulate the developmental Program of the early human placenta. Stem Cell Rep 2018;11:537–51. https://doi.org/10.1016/j.stemcr.2018.07.004.Search in Google Scholar PubMed PubMed Central
31. Park, JY, Mani, S, Clair, G, Olson, HM, Paurus, VL, Ansong, CK, et al.. A microphysiological model of human trophoblast invasion during implantation. Nat Commun 2022;13:1252. https://doi.org/10.1038/s41467-022-28663-4.Search in Google Scholar PubMed PubMed Central
32. Bedzhov, I, Zernicka-Goetz, M. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell 2014;156:1032–44. https://doi.org/10.1016/j.cell.2014.01.023.Search in Google Scholar PubMed PubMed Central
33. Bedzhov, I, Leung, CY, Bialecka, M, Zernicka-Goetz, M. In vitro culture of mouse blastocysts beyond the implantation stages. Nat Protoc 2014;9:2732–9. https://doi.org/10.1038/nprot.2014.186.Search in Google Scholar PubMed
34. Taniguchi, K, Shao, Y, Townshend, RF, Tsai, YH, DeLong, CJ, Lopez, SA, et al.. Lumen formation is an intrinsic property of isolated human pluripotent stem cells. Stem Cell Rep 2015;5:954–62. https://doi.org/10.1016/j.stemcr.2015.10.015.Search in Google Scholar PubMed PubMed Central
35. Shao, Y, Taniguchi, K, Gurdziel, K, Townshend, RF, Xue, X, Yong, KMA, et al.. Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat Mater 2017;16:419–25. https://doi.org/10.1038/nmat4829.Search in Google Scholar PubMed PubMed Central
36. Zheng, Y, Xue, X, Shao, Y, Wang, S, Esfahani, SN, Li, Z, et al.. Controlled modelling of human epiblast and amnion development using stem cells. Nature 2019;573:421–5. https://doi.org/10.1038/s41586-019-1535-2.Search in Google Scholar PubMed PubMed Central
37. Warmflash, A, Sorre, B, Etoc, F, Siggia, ED, Brivanlou, AH. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods 2014;11:847–54. https://doi.org/10.1038/nmeth.3016.Search in Google Scholar PubMed PubMed Central
38. Martyn, I, Kanno, TY, Ruzo, A, Siggia, ED, Brivanlou, AH. Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature 2018;558:132–5. https://doi.org/10.1038/s41586-018-0150-y.Search in Google Scholar PubMed PubMed Central
39. Simunovic, M, Metzger, JJ, Etoc, F, Yoney, A, Ruzo, A, Martyn, I, et al.. A 3D model of a human epiblast reveals BMP4-driven symmetry breaking. Nat Cell Biol 2019;21:900–10. https://doi.org/10.1038/s41556-019-0349-7.Search in Google Scholar PubMed
40. van den Brink, SC, Baillie-Johnson, P, Balayo, T, Hadjantonakis, AK, Nowotschin, S, Turner, DA, et al.. Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 2014;141:4231–42. https://doi.org/10.1242/dev.113001.Search in Google Scholar PubMed PubMed Central
41. Beccari, L, Moris, N, Girgin, M, Turner, DA, Baillie-Johnson, P, Cossy, AC, et al.. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 2018;562:272–6. https://doi.org/10.1038/s41586-018-0578-0.Search in Google Scholar PubMed
42. Moris, N, Anlas, K, van den Brink, SC, Alemany, A, Schröder, J, Ghimire, S, et al.. An in vitro model of early anteroposterior organization during human development. Nature 2020;582:410–5. https://doi.org/10.1038/s41586-020-2383-9.Search in Google Scholar PubMed
43. van den Brink, SC, Alemany, A, van Batenburg, V, Moris, N, Blotenburg, M, Vivié, J, et al.. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 2020;582:405–9. https://doi.org/10.1038/s41586-020-2024-3.Search in Google Scholar PubMed
44. Wen, L, Tang, F. Single-cell sequencing in stem cell biology. Genome Biol 2016;17:71. https://doi.org/10.1186/s13059-016-0941-0.Search in Google Scholar PubMed PubMed Central
45. Haniffa, M, Taylor, D, Linnarsson, S, Aronow, BJ, Bader, GD, Barker, RA, et al.. A roadmap for the human developmental cell atlas. Nature 2021;597:196–205. https://doi.org/10.1038/s41586-021-03620-1.Search in Google Scholar PubMed
46. Yan, L, Yang, M, Guo, H, Yang, L, Wu, J, Li, R, et al.. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 2013;20:1131–9. https://doi.org/10.1038/nsmb.2660.Search in Google Scholar PubMed
47. Petropoulos, S, Edsgärd, D, Reinius, B, Deng, Q, Panula, SP, Codeluppi, S, et al.. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 2016;165:1012–26. https://doi.org/10.1016/j.cell.2016.03.023.Search in Google Scholar PubMed PubMed Central
48. Zhou, F, Wang, R, Yuan, P, Ren, Y, Mao, Y, Li, R, et al.. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature 2019;572:660–4. https://doi.org/10.1038/s41586-019-1500-0.Search in Google Scholar PubMed
49. Xiang, L, Yin, Y, Zheng, Y, Ma, Y, Li, Y, Zhao, Z, et al.. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 2020;577:537–42. https://doi.org/10.1038/s41586-019-1875-y.Search in Google Scholar PubMed
50. Tyser, RCV, Mahammadov, E, Nakanoh, S, Vallier, L, Scialdone, A, Srinivas, S. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature 2021;600:285–9. https://doi.org/10.1038/s41586-021-04158-y.Search in Google Scholar PubMed
51. Nakamura, T, Okamoto, I, Sasaki, K, Yabuta, Y, Iwatani, C, Tsuchiya, H, et al.. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 2016;537:57–62. https://doi.org/10.1038/nature19096.Search in Google Scholar PubMed
52. Niu, Y, Sun, N, Li, C, Lei, Y, Huang, Z, Wu, J, et al.. Dissecting primate early post-implantation development using long-term in vitro embryo culture. Science 2019;366:eaaw5754. https://doi.org/10.1126/science.aaw5754.Search in Google Scholar PubMed
53. Ma, H, Zhai, J, Wan, H, Jiang, X, Wang, X, Wang, L, et al.. In vitro culture of cynomolgus monkey embryos beyond early gastrulation. Science 2019;366:eaax7890. https://doi.org/10.1126/science.aax7890.Search in Google Scholar PubMed
54. Grün, D, Lyubimova, A, Kester, L, Wiebrands, K, Basak, O, Sasaki, N, et al.. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 2015;525:251–5. https://doi.org/10.1038/nature14966.Search in Google Scholar PubMed
55. Wen, L, Tang, F. Recent advances in single-cell sequencing technologies. Precis Clin Med 2022;5:pbac002. https://doi.org/10.1093/pcmedi/pbac002.Search in Google Scholar PubMed PubMed Central
56. Zhang, K, Hocker, JD, Miller, M, Hou, X, Chiou, J, Poirion, OB, et al.. A single-cell atlas of chromatin accessibility in the human genome. Cell 2021;184:5985–6001.e19. https://doi.org/10.1016/j.cell.2021.10.024.Search in Google Scholar PubMed PubMed Central
57. Asp, M, Bergenstråhle, J, Lundeberg, J. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays 2020;42:e1900221. https://doi.org/10.1002/bies.201900221.Search in Google Scholar PubMed
58. Li, G, Li, X, Zhuang, S, Wang, L, Zhu, Y, Chen, Y, et al.. Gene editing and its applications in biomedicine. Sci China Life Sci 2022;65:660–700. https://doi.org/10.1007/s11427-021-2057-0.Search in Google Scholar PubMed PubMed Central
59. Esk, C, Lindenhofer, D, Haendeler, S, Wester, RA, Pflug, F, Schroeder, B, et al.. A human tissue screen identifies a regulator of ER secretion as a brain-size determinant. Science 2020;370:935–41. https://doi.org/10.1126/science.abb5390.Search in Google Scholar PubMed
60. Song, Y, van den Berg, PR, Markoulaki, S, Soldner, F, Dall’Agnese, A, Henninger, JE, et al.. Dynamic enhancer DNA methylation as basis for transcriptional and cellular heterogeneity of ESCs. Mol Cell 2019;75:905–20.e6. https://doi.org/10.1016/j.molcel.2019.06.045.Search in Google Scholar PubMed PubMed Central
61. Agoglia, RM, Sun, D, Birey, F, Yoon, SJ, Miura, Y, Sabatini, K, et al.. Primate cell fusion disentangles gene regulatory divergence in neurodevelopment. Nature 2021;592:421–7. https://doi.org/10.1038/s41586-021-03343-3.Search in Google Scholar PubMed PubMed Central
62. Ju, YS, Martincorena, I, Gerstung, M, Petljak, M, Alexandrov, LB, Rahbari, R, et al.. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature 2017;543:714–8. https://doi.org/10.1038/nature21703.Search in Google Scholar PubMed PubMed Central
63. Coorens, THH, Moore, L, Robinson, PS, Sanghvi, R, Christopher, J, Hewinson, J, et al.. Extensive phylogenies of human development inferred from somatic mutations. Nature 2021;597:387–92. https://doi.org/10.1038/s41586-021-03790-y.Search in Google Scholar PubMed
64. Sakaue-Sawano, A, Yo, M, Komatsu, N, Hiratsuka, T, Kogure, T, Hoshida, T, et al.. Genetically encoded tools for optical dissection of the mammalian cell cycle. Mol Cell 2017;68:626–40.e5. https://doi.org/10.1016/j.molcel.2017.10.001.Search in Google Scholar PubMed
65. Shimokawa, M, Ohta, Y, Nishikori, S, Matano, M, Takano, A, Fujii, M, et al.. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature 2017;545:187–92. https://doi.org/10.1038/nature22081.Search in Google Scholar PubMed
66. Ohta, Y, Fujii, M, Takahashi, S, Takano, A, Nanki, K, Matano, M, et al.. Cell-matrix interface regulates dormancy in human colon cancer stem cells. Nature 2022;608:784–94. https://doi.org/10.1038/s41586-022-05043-y.Search in Google Scholar PubMed
67. Kalhor, R, Kalhor, K, Mejia, L, Leeper, K, Graveline, A, Mali, P, et al.. Developmental barcoding of whole mouse via homing CRISPR. Science 2018;361:eaat9804. https://doi.org/10.1126/science.aat9804.Search in Google Scholar PubMed PubMed Central
68. Chan, MM, Smith, ZD, Grosswendt, S, Kretzmer, H, Norman, TM, Adamson, B, et al.. Molecular recording of mammalian embryogenesis. Nature 2019;570:77–82. https://doi.org/10.1038/s41586-019-1184-5.Search in Google Scholar PubMed PubMed Central
69. Qian, X, Nguyen, HN, Song, MM, Hadiono, C, Ogden, SC, Hammack, C, et al.. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 2016;165:1238–54. https://doi.org/10.1016/j.cell.2016.04.032.Search in Google Scholar PubMed PubMed Central
70. Aguilera-Castrejon, A, Oldak, B, Shani, T, Ghanem, N, Itzkovich, C, Slomovich, S, et al.. Ex utero mouse embryogenesis from pre-gastrulation to late organogenesis. Nature 2021;593:119–24. https://doi.org/10.1038/s41586-021-03416-3.Search in Google Scholar PubMed
71. Simunovic, M, Brivanlou, AH. Embryoids, organoids and gastruloids: new approaches to understanding embryogenesis. Development 2017;144:976–85. https://doi.org/10.1242/dev.143529.Search in Google Scholar PubMed PubMed Central
72. Lancaster, MA, Corsini, NS, Wolfinger, S, Gustafson, EH, Phillips, AW, Burkard, TR, et al.. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol 2017;35:659–66. https://doi.org/10.1038/nbt.3906.Search in Google Scholar PubMed PubMed Central
73. Cederquist, GY, Asciolla, JJ, Tchieu, J, Walsh, RM, Cornacchia, D, Resh, MD, et al.. Specification of positional identity in forebrain organoids. Nat Biotechnol 2019;37:436–44. https://doi.org/10.1038/s41587-019-0085-3.Search in Google Scholar PubMed PubMed Central
74. Birey, F, Andersen, J, Makinson, CD, Islam, S, Wei, W, Huber, N, et al.. Assembly of functionally integrated human forebrain spheroids. Nature 2017;545:54–9. https://doi.org/10.1038/nature22330.Search in Google Scholar PubMed PubMed Central
75. Workman, MJ, Mahe, MM, Trisno, S, Poling, HM, Watson, CL, Sundaram, N, et al.. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med 2017;23:49–59. https://doi.org/10.1038/nm.4233.Search in Google Scholar PubMed PubMed Central
76. Taguchi, A, Nishinakamura, R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell 2017;21:730–46.e6. https://doi.org/10.1016/j.stem.2017.10.011.Search in Google Scholar PubMed
77. Murphy, SV, Atala, A. 3D bioprinting of tissues and organs. Nat Biotechnol 2014;32:773–85. https://doi.org/10.1038/nbt.2958.Search in Google Scholar PubMed
78. Ren, Y, Yang, X, Ma, Z, Sun, X, Zhang, Y, Li, W, et al.. Developments and opportunities for 3D bioprinted organoids. Int J Bioprint 2021;7:364. https://doi.org/10.18063/ijb.v7i3.364.Search in Google Scholar PubMed PubMed Central
79. Zhang, Z, Wu, C, Dai, C, Shi, Q, Fang, G, Xie, D, et al.. A multi-axis robot-based bioprinting system supporting natural cell function preservation and cardiac tissue fabrication. Bioact Mater 2022;18:138–50. https://doi.org/10.1016/j.bioactmat.2022.02.009.Search in Google Scholar PubMed PubMed Central
80. Kupfer, ME, Lin, WH, Ravikumar, V, Qiu, K, Wang, L, Gao, L, et al.. In situ expansion, differentiation, and electromechanical coupling of human cardiac muscle in a 3D bioprinted, chambered organoid. Circ Res 2020;127:207–24. https://doi.org/10.1161/circresaha.119.316155.Search in Google Scholar
81. Brassard, JA, Nikolaev, M, Hübscher, T, Hofer, M, Lutolf, MP. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat Mater 2021;20:22–9. https://doi.org/10.1038/s41563-020-00803-5.Search in Google Scholar PubMed
82. Lee, A, Hudson, AR, Shiwarski, DJ, Tashman, JW, Hinton, TJ, Yerneni, S, et al.. 3D bioprinting of collagen to rebuild components of the human heart. Science 2019;365:482–7. https://doi.org/10.1126/science.aav9051.Search in Google Scholar PubMed
83. Ren, X, Moser, PT, Gilpin, SE, Okamoto, T, Wu, T, Tapias, LF, et al.. Engineering pulmonary vasculature in decellularized rat and human lungs. Nat Biotechnol 2015;33:1097–102. https://doi.org/10.1038/nbt.3354.Search in Google Scholar PubMed
84. Koot, YE, Teklenburg, G, Salker, MS, Brosens, JJ, Macklon, NS. Molecular aspects of implantation failure. Biochim Biophys Acta 2012;1822:1943–50. https://doi.org/10.1016/j.bbadis.2012.05.017.Search in Google Scholar PubMed
85. Li, R, Sun, L, Fang, A, Li, P, Wu, Q, Wang, X. Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell 2017;8:823–33. https://doi.org/10.1007/s13238-017-0479-2.Search in Google Scholar PubMed PubMed Central
86. Zhao, B, Ni, C, Gao, R, Wang, Y, Yang, L, Wei, J, et al.. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell 2020;11:771–5. https://doi.org/10.1007/s13238-020-00718-6.Search in Google Scholar PubMed PubMed Central
87. Gao, Y, Zhang, J, Liu, Z, Qi, S, Guo, X, Wang, H, et al.. Single-cell sequencing reveals clearance of blastula chromosomal mosaicism in in Vitro fertilization babies. Dev Reprod Biol 2022;S1672–0229:00088–2. https://doi.org/10.1016/j.gpb.2022.07.004.Search in Google Scholar PubMed
88. Yang, M, Rito, T, Metzger, J, Naftaly, J, Soman, R, Hu, J, et al.. Depletion of aneuploid cells in human embryos and gastruloids. Nat Cell Biol 2021;23:314–21. https://doi.org/10.1038/s41556-021-00660-7.Search in Google Scholar PubMed
89. Paulsen, B, Velasco, S, Kedaigle, AJ, Pigoni, M, Quadrato, G, Deo, AJ, et al.. Autism genes converge on asynchronous development of shared neuron classes. Nature 2022;602:268–73. https://doi.org/10.1038/s41586-021-04358-6.Search in Google Scholar PubMed PubMed Central
90. Kelley, KW, Pasca, SP. Human brain organogenesis: toward a cellular understanding of development and disease. Cell 2022;185:42–61. https://doi.org/10.1016/j.cell.2021.10.003.Search in Google Scholar PubMed
91. Hu, W, Jiang, C, Guan, D, Dierickx, P, Zhang, R, Moscati, A, et al.. Patient Adipose stem cell-derived adipocytes reveal genetic variation that predicts antidiabetic drug response. Cell Stem Cell 2019;24:299–308.e6. https://doi.org/10.1016/j.stem.2018.11.018.Search in Google Scholar PubMed PubMed Central
92. Tuveson, D, Clevers, H. Cancer modeling meets human organoid technology. Science 2019;364:952–5. https://doi.org/10.1126/science.aaw6985.Search in Google Scholar PubMed
93. Wang, R, Mao, Y, Wang, W, Zhou, X, Wang, W, Gao, S, et al.. Systematic evaluation of colorectal cancer organoid system by single-cell RNA-Seq analysis. Genome Biol 2022;23:106. https://doi.org/10.1186/s13059-022-02673-3.Search in Google Scholar PubMed PubMed Central
94. Neal, JT, Li, X, Zhu, J, Giangarra, V, Grzeskowiak, CL, Ju, J, et al.. Organoid modeling of the tumor immune microenvironment. Cell 2018;175:1972–88.e16. https://doi.org/10.1016/j.cell.2018.11.021.Search in Google Scholar PubMed PubMed Central
95. Bian, S, Repic, M, Guo, Z, Kavirayani, A, Burkard, T, Bagley, JA, et al.. Genetically engineered cerebral organoids model brain tumor formation. Nat Methods 2018;15:631–9. https://doi.org/10.1038/s41592-018-0070-7.Search in Google Scholar PubMed PubMed Central
Supplementary Material
The online version of this article offers supplementary material (https://doi.org/10.1515/mr-2022-0028).
© 2022 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorial
- Exploring the mysteries of reproductive health
- Reviews
- Developmental origins of adult diseases
- Site-specific genome editing in treatment of inherited diseases: possibility, progress, and perspectives
- An update on placental drug transport and its relevance to fetal drug exposure
- Organoid research on human early development and beyond
- Fibroblast growth factor 21 and dietary interventions: what we know and what we need to know next
- Hypothalamic GABAergic neurocircuitry in the regulation of energy homeostasis and sleep/wake control
Articles in the same Issue
- Frontmatter
- Editorial
- Exploring the mysteries of reproductive health
- Reviews
- Developmental origins of adult diseases
- Site-specific genome editing in treatment of inherited diseases: possibility, progress, and perspectives
- An update on placental drug transport and its relevance to fetal drug exposure
- Organoid research on human early development and beyond
- Fibroblast growth factor 21 and dietary interventions: what we know and what we need to know next
- Hypothalamic GABAergic neurocircuitry in the regulation of energy homeostasis and sleep/wake control

