Assessment of long-term stored specimens in the Siriraj Hospital colorectal cancer biobank for RNA sequencing and profiling
-
Thanawat Suwatthanarak
and Vitoon Chinswangwatanakul
Abstract
Objectives
Biobanks play an important role in advancing cancer research, yet concerns persist regarding the molecular integrity of long-term stored samples. This study assessed fresh frozen (FF) tissues and formalin-fixed paraffin-embedded (FFPE) tissues from the Siriraj Hospital colorectal cancer (CRC) biobank collected during two distinct periods (2011–2012 and 2020–2021).
Methods
In 2022, FF and FFPE primary cancer tissues from 75 CRC patients were evaluated. RNA sequencing (RNA-Seq) analyzed comprehensive gene expression profiles in FF tissues preserved at −80 °C, while nCounter profiling elucidated cancer-specific RNA transcripts in FFPE tissues stored at ambient temperature. Comparative analyses were conducted between specimens from 2011 to 2012 and 2020–2021.
Results
The FF tissues stored for approximately 10.5 years were well-suited for RNA-Seq compared to the intact tissues preserved for 1.5 years. Despite consistencies in RNA quantity, RNA integrity, amount of sequencing reads, and CRC gene signature, gene enrichment analysis revealed the decreased ribosome biogenesis, spliceosome and antifolate resistance pathways in the 2011–2012 group. Moreover, the FFPE tissues also showed no alteration in RNA quantity between the two periods, and the nCounter profiling demonstrated comparable CRC-specific gene counts in spite of the significant reduction of raw counts in the 2011–2012 group.
Conclusions
We report that FF tissues from CRC patients, stored for 10 years, are viable for whole transcriptome RNA-Seq, despite altered pathways such as ribosome biogenesis, spliceosome, and antifolate resistance. Moreover, 10-year-stored FFPE CRC tissues remain suitable for specific RNA profiling using the nCounter pan-cancer panel, despite a significant reduction in raw counts. These findings underscore the enduring contribution of biobanks to molecular research, highlighting their value a decade post-collection.
Introduction
Colorectal cancer (CRC) represents a significant global health challenge that requires continuous advancements in diagnostic and therapeutic strategies [1]. Biobanks, repositories of well-preserved biological specimens, play a pivotal role in facilitating cutting-edge research by providing access to diverse biological materials for molecular analyzes [2]. These repositories harbor a wealth of specimens collected over various time frames and stored through different preservation methods [3]. One such repository is the CRC biobank at Siriraj Hospital, which houses a collection of specimens preserved as fresh frozen (FF) and formalin-fixed, paraffin-embedded (FFPE) tissues.
While biobanks offer invaluable resources, the integrity and utility of long-term stored specimens remain critical considerations, especially for studies that require molecular characterization [4]. Over time, concerns about RNA degradation and alterations in gene integrity and expression, can potentially affect the reliability and relevance of results obtained from these specimens [5].
Recently, advanced molecular techniques have revolutionized our ability to unravel the intricate complexities of biological systems, particularly in the realm of cancer research [6]. Among all techniques, RNA sequencing (RNA-Seq) and nCounter RNA profiling stand out as transformative tools that have reshaped our understanding of cellular processes, disease mechanisms, and therapeutic interventions [7, 8]. RNA-Seq offers a comprehensive view of gene expression patterns by quantifying RNA molecules, allowing us to identify key players in cancer progression and uncover novel regulatory elements [7]. nCounter RNA profiling, with its digital barcoding system, facilitates precise quantification of target transcripts, allowing the validation of biomarkers and the elucidation of intricate gene networks [8]. These two techniques collectively propel cancer research into a new era of precision medicine, fostering insights that not only expand our fundamental understanding but also drive the development of innovative therapeutic strategies [7, 8].
This study seeks to address concerns by performing an assessment of long-term stored FF and FFPE specimens in the Siriraj Hospital CRC biobank. Using two molecular techniques, including RNA-Seq and nCounter RNA profiling, this investigation aims to determine and compare the gene expression patterns and signatures of CRC specimens collected during two distinct timeframes: 2011–2012 and 2020–2021. The specimens collected in 2020–2021, stored for approximately 1.5 years, and known to retain molecular integrity, were comparatively used as the reference standard [9, 10]. We seek to inform best practices for using biobank resources in future molecular investigations.
Materials and methods
Participating patients and ethical considerations
Patients at least 18 years of age who were diagnosed with colorectal adenocarcinoma at Siriraj Hospital, Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand, were included in this study. All participants were treated according to the National Comprehensive Cancer Network guidelines. This study was part of a research series approved by the Siriraj Institutional Review Board (COA No. Si156/2011, Si348/2019 and Si105/2021). Ethical considerations and informed consent have been followed during the collection of FF and FFPE tissues from a total of 75 participating individuals during two distinct timeframes, 2011–2012 and 2020–2021. Table 1 shows the demographic data of the participating patients.
Demographic data of the participants.
| Characteristics | 2011–2012 | 2020–2021 | p-Valuea |
|---|---|---|---|
| Sex Male Female |
27 (52.9 %) 24 (47.1 %) |
16 (66.7 %) 8 (33.3 %) |
0.262 |
| Age Range, years Mean ± standard deviation, years |
30 to 86 63.12 ± 12.18 |
49 to 75 62.96 ± 9.45 |
0.951 |
| Tumor location Left-sided colon Right-sided colon Rectum |
24 (41.2 %) 13 (25.5 %) 14 (33.3 %) |
12 (50.0 %) 6 (25.0 %) 6 (25.0 %) |
0.967 |
| Stage I-II III-IV |
10 (19.6 %) 41 (80.4 %) |
0 (0.0 %) 24 (100.0 %) |
0.025 |
| Pathology grade Well differentiated Moderately differentiated Poorly differentiated |
2 (3.9 %) 48 (94.1 %) 1 (2.0 %) |
1 (4.2 %) 23 (95.8 %) 0 (0.0 %) |
0.787 |
-
aThe p-value was calculated using the T-test, Chi-square, or Fisher’s exact test.
Sample collection and storage
For FF tissues, within 30 min after surgical resection, the freshly obtained primary CRC tissues were promptly cut into 2–3 mm pieces. They were immediately washed with phosphate-buffered saline (pH ∼ 7.4) and then immersed in 1 mL of chilled RNAlater solution (Invitrogen, MA, USA) on ice, following the protocol outlined in our previous report [11]. Subsequently, the specimens were stored at −80 °C in the Siriraj CRC biobank until they were required for further use.
To prepare FFPE tissues, the resected primary CRC tissues were fixed in 10 % neutral buffered formalin within 60 min after surgical resection. The fixation process lasted for 90 min. Subsequently, the fixed tissues underwent dehydration using ascending grades of ethyl alcohol, including 95 % ethanol for 30 min (2 times) and absolute ethanol for 60 min (3 times). Following dehydration, the tissues underwent clearing by immersion in xylene for 60 min (2 times). For infiltration, the dehydrated and cleared tissues were then immersed in molten paraffin wax (maintained at 60–64 °C) for 60 min (3 times). After infiltration, the tissues were oriented in molds filled with molten paraffin wax (maintained at 60–65 °C) and allowed to solidify on a cold plate set at −5 °C. Finally, the resulting paraffin blocks containing the embedded tissue were stored at ambient temperature until further use.
Notably, all specimens were collected by surgeons and assessed by pathologists. The tissue collection, preparation, and storage protocols remained consistent across two distinct periods, spanning from 2011 to 2012 and from 2020 to 2021. All specimens were stored until all experiments were conducted in 2022. Therefore, the storage time for the specimens collected 2011–2012 and 2020–2021 was approximately 10.5 and 1.5 years, respectively. The difference in storage time between the two groups was approximately 9 years.
RNA-Seq
Total RNA was extracted from the stored FF tissues. A lysis buffer together with the lysing matrix Z (MP Biomedicals, Santa Ana, CA, USA) was applied to each FF tissue (30 mg) before homogenization using a FastPrep-24 5G instrument (MP Biomedicals, CA, USA) at a speed of 6.0 m/s for 40 s on and 3 min off cycles. The homogenization step was continued until a clear lysate was obtained. The RNeasy mini kit (Qiagen, Hilden, Germany) was used to extract total RNA according to the manufacturer’s instructions. Approximately 30 μL RNA eluates were obtained. Concentration and RNA integrity number (RIN) were assessed using the 5,400 fragment analyzer system (Agilent Technologies, CA, USA).
RNA library preparation and sequencing were performed by Novogene Co. Ltd. (Singapore) using human strand-specific mRNA (WBI-Quantification), directional mRNA library preparation (poly A enrichment) and 2 × 150 bp pair-end configuration (PE) with raw data of 8.0 Gb per sample. Briefly, isolation of mRNA from total RNA was achieved by utilizing magnetic beads coupled with poly-T oligos. Following fragmentation, the first strand of cDNA was synthesized via random hexamer primers, followed by the subsequent synthesis of the second cDNA strand using dUTP for directional library. The library’s quality assessment was conducted using Qubit and real-time polymerase chain reaction for quantification, as well as a bioanalyzer for size distribution analysis. The prepared libraries were subsequently pooled for sequencing on the NovaSeq platform (Illumina, San Diego, CA, USA), according to optimal library concentration and data amount. All samples were sequenced in the same batch. During the data quality control process, raw reads of the FASTQ format were processed before clean reads were obtained for downstream analyzes. We conducted quantification of gene expression levels, co-expression analysis, and enrichment analysis based on whole transcriptome RNA-Seq data. Cell deconvolution in each FF sample was calculated by python-based cellanneal software using the RNA-Seq data and using the signature of human primary cells from the human cell atlas [12].
nCounter RNA profiling
Total RNA was extracted from the stored FFPE tissues (two sections with 5 μm thickness) using a high purity FFPE RNA isolation kit (Roche Diagnostics, Indianapolis, IND, USA), according to the manufacturer’s instructions. Approximately 30 μL RNA eluates were obtained and the RNA concentration was analyzed using a Nanodrop 8,000 spectrophotometer (Thermo Fisher Scientific, MA, USA). Besides, the 18S fragment size was analyzed by the Agilent 2,100 Bioanalyzer and RNA 6000 Nano Chip (Agilent Technologies, CA, USA).
Gene expression profiling was performed using the nCounter PanCancer Progression Panel, which encompasses 770 cancer-associated genes (NanoString Technologies, WA, USA). Each sample, comprising 300 ng of RNA, was hybridized involving capture probes and reporter probes via CodeSet hybridization, conducted at 65 °C for a duration of 18 h. Subsequently, the hybridized specimens were processed on the automated nCounter Prep Station (NanoString Technologies, WA, USA), and the nCounter cartridges obtained were interpreted using the nCounter Digital Analyzer (NanoString Technologies, WA, USA). Data processing was performed with the nSolver Analysis Software version 4.0 (NanoString Technologies, WA, USA), with raw counts being normalized against the geometric mean counts of 11 housekeeping genes (HRNP1, RPL27, RPL9, RPL6, RPL30, OAZ1, PTMA, RPS29, UBC, RPS12, and RPS16). Cell deconvolution for each FFPE sample was determined using python-based cellanneal software, the nCounter RNA data, and the signature of human primary cells from the human cell atlas [12].
Statistical analysis
IBM SPSS Statistics (SPSS Inc., IL, USA) or Prism (GraphPad Software, Inc., CA, USA) was used to perform statistical analyzes using appropriate methods, such as the T test, the Mann-Whitney U test, the Chi-square test, and Fisher’s exact test to determine differences between the 2011–2012 and 2020–2021 groups. A p-value of <0.05 was considered statistically significant.
Results
Assessment of long-term stored FF specimens from CRC patients for RNA-Seq
In this section, we conducted an examination of FF CRC primary tissues collected during two distinct time periods, 2011–2012 and 2020–2021, with the objective of evaluating their molecular integrity and suitability for RNA-Seq, a high-throughput technique critical for understanding gene expression patterns and molecular mechanisms in cancer [13]. Our analysis involved 30 FF tissue specimens collected from a cohort of 30 CRC patients (Figure 1A). The tissues collected during the two specified time frames, which had an approximate 9-year difference in storage time, were subjected to RNA extraction and subsequent RNA-Seq to assess their suitability for downstream molecular analyzes.

Assessment of long-term stored FF specimens from CRC patients for RNA-Seq. (A) Patient participation chart. (B) RNA concentration. (C) RIN value. (D) Raw reads. (E) Clean reads. (F) Tumor cell fraction. p-Value (p) was calculated using the Mann-Whitney U test.
First, the quality control processes of the samples and sequencing data were focused. As shown in Figure 1B and C, the RNA extracted from both time frames exhibited consistent concentration and integrity. There were no significant differences in concentration and RIN values between both groups, indicating optimal RNA preservation, despite the extended storage durations (Figure 1B and C). Furthermore, in terms of sequencing data quality, the data obtained from both sample groups were consistent in raw and clean reads (Figure 1D and E). From cell deconvolution, FF tissues contained tumor cells (colorectal adenocarcinoma) with an average of approximately 28 % (Supplementary Figure S1 and Figure 1F). There was no significant difference of tumor cell content in both groups of FF tissues (Figure 1F).
In addition to quality control processes, CRC-specific gene signature, coexpression, and enrichment analyzes were performed. The expression of four genes (BDNF, CTNNB1, GSK3B and PTGS2), which were reported to be upregulated in primary tumor tissues of CRC compared to adjacent normal tissues [14] was analyzed. Interestingly, comparative analysis between the two timeframes demonstrated a concordance in the four-gene CRC signature (Figure 2A). Figure 2B shows the Venn coexpression diagram, indicating the number of genes that are uniquely expressed and co-expressed in the two groups. Both groups co-expressed approximately 93 % of the genes analyzed (Figure 2B). As a result of enrichment analysis by the collection of Kyoto Encyclopedia of Genes and Genomes (KEGG), ribosome biogenesis, spliceosome and antifolate resistance pathways were significantly altered in the observed gene expression patterns of the two groups (Figure 2B).
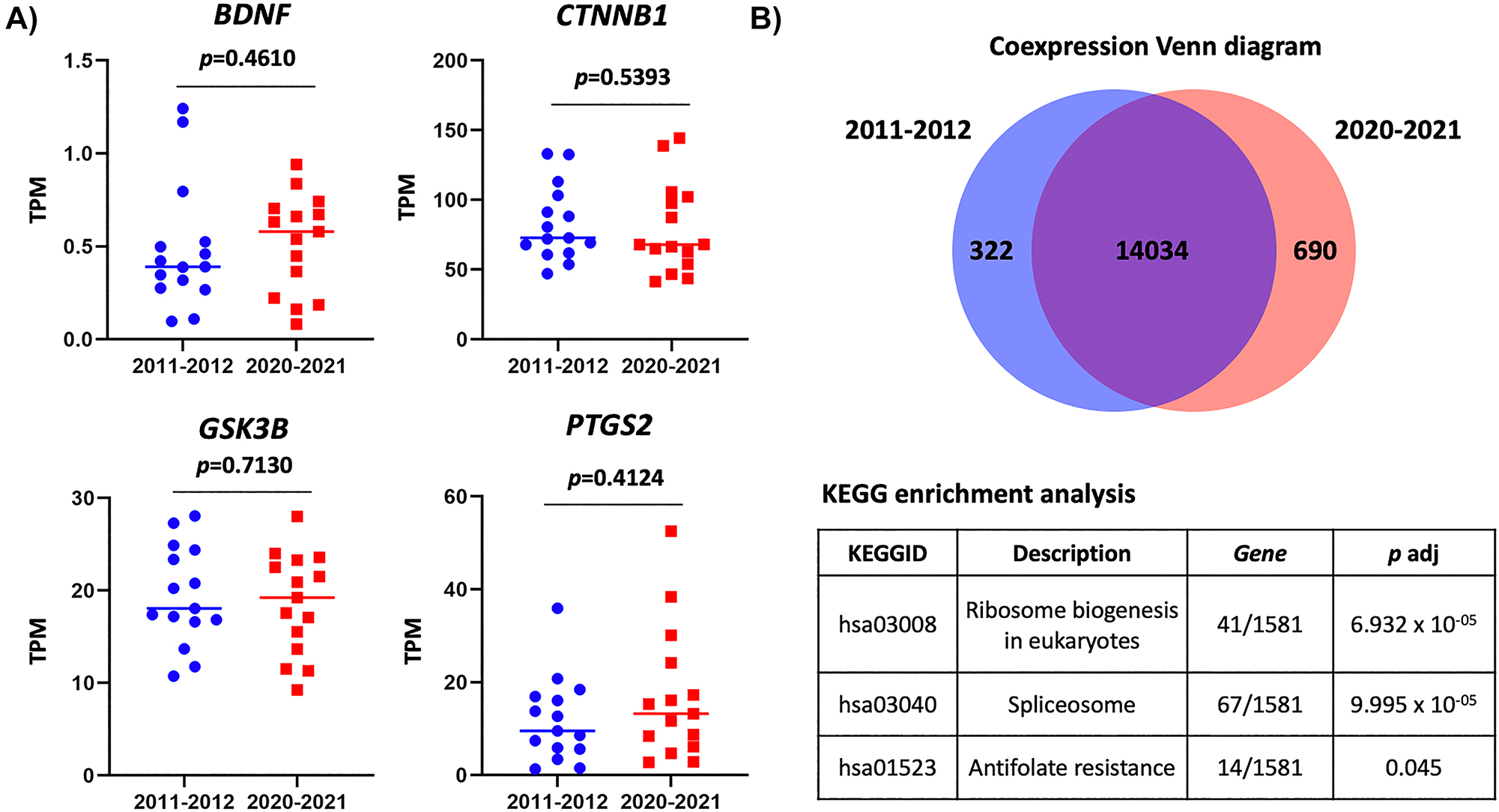
Analysis of RNA-Seq data from long-term stored FF CRC specimens. (A) Transcript per million value of four CRC-signature genes. p-Value (p) was calculated using the Mann-Whitney U test. (B) Venn diagram of coexpression analysis (top) and table of KEGG: Kyoto Encyclopedia of Genes and Genomes enrichment analysis (bottom).
This investigation underscores the remarkable molecular suitability of long-term stored FF CRC primary tissues. Despite storage over an extended period, these specimens retain RNA suitable for RNA-Seq analyzes. Consistent concentration, integrity, sequencing read generation, gene signature expression, and gene coexpression of the two groups affirmed the suitability of approximately 10-year-stored FF primary tissues for exploring CRC biology. However, the pathways related to ribosome biogenesis in eukaryotes, spliceosome, and antifolate resistance should be noted when the samples were collected in the different periods. This finding further emphasizes the lasting value of biobank resources to advance oncological research.
Assessment of long-term stored FFPE specimens from CRC patients for RNA nCounter profiling
For this section, we performed an assessment of FFPE CRC primary tissues collected during two distinct time periods, 2011–2012 and 2020–2021. Our objective was to evaluate the molecular suitability of long-term stored FFPE tissues for nCounter RNA profiling, a promising technique for quantifying specific RNA transcripts and elucidating gene expression patterns in cancer [15]. The study involved 58 FFPE CRC primary tissues (36 tissues in 2011–2012 and 22 tissues in 2020–2021) collected from a cohort of 45 CRC patients (Figure 3A). In some patients, two or more archived FFPE blocks were used (Figure 3A). These FFPE tissues were processed over specified timeframes to extract RNA for subsequent nCounter RNA profiling, with the aim of assessing their suitability for CRC-related molecular analysis.

Assessment of long-term stored FFPE specimens from CRC patients for RNA nCounter profiling. (A) Patient participation chart. (B) RNA concentration. (C) 18S fragment size. (D) Total raw counts. (E) Total normalized counts. (F) Tumor cell fraction. p-Value (p) was calculated using the T-test or Mann-Whitney U test.
Despite prolonged storage periods, the RNA extracted from both time frames did not show significant differences in yield or concentration (Figure 3B). However, the reduction of 18S fragment size in the older samples was observed (Figure 3C). Following the hybridization and counting of 770 target genes in the Nanostring PanCancer panel, gene counts and CRC-specific genes were analyzed. The results of the RNA nCounter profiling showed that the FFPE tissues from 2011 to 2012 significantly produced lower raw counts than the FFPE tissues from 2020 to 2021 (Figure 3D). However, the total of normalized counts was comparable between the two groups after normalization by the housekeeping genes (Figure 3E). Based on cell deconvolution analysis, FFPE tissues showed an average tumor cell content of approximately 22 % (Supplementary Figure S2 and Figure 3F). No significant difference in tumor cell content was observed between the two groups of FFPE tissues (Figure 3F).
Five genes, including COL1A1, COMP, ETV4, SPP1 and INHBA, were then analyzed, since they were included in the Nanostring PanCancer panel and reported to be differentially expressed in tissues of colon adenocarcinoma versus normal tissues of the colon [16]. The results of the nCounter RNA profiling also demonstrated that the FFPE tissues from both time periods produced comparable normalized counts of these CRC-specific genes (Figure 4). These 5 CRC-related genes also did not exhibit significant differences between the two storage periods in FF tissues (Supplementary Figure S3). This comparative analysis between 2011-2012 and 2020–2021 specimens revealed consistent normalized expression profiles of these CRC-specific genes, reaffirming the utility of approximately 10-year-stored FFPE primary tissues for investigating gene expression patterns in CRC.

Normalized counts for five CRC-signature genes in the PanCancer progression panel. p-Value (p) was calculated using the Mann-Whitney U test.
Discussion
The evaluation of long-term stored specimens in the Siriraj Hospital CRC biobank presents a comparative exploration of the molecular integrity and utility of archived primary CRC FF and FFPE tissues from two different periods (2011–2012 and 2020–2021) for two molecular analyzes, including RNA-Seq and nCounter RNA profiling.
Our finding highlights the molecular stability observed in FF tissues preserved for approximately a decade in RNAlater, an RNA stabilization solution. Despite prolonged storage durations, the RNA extracted from FF samples from both storage periods consistently exhibited significant quantity and integrity. This stability underscores the reliability of FF tissues in RNAlater as a valuable resource for transcriptomic analyzes and supports the continued use of biobanked FF specimens to investigate CRC biology.
Equally noteworthy is the enduring value of FFPE tissues, preserved through formalin fixation and paraffin embedding. These tissues, often collected as part of routine clinical practice, have the advantage of being available for retrospective studies. Importantly, RNA extracted from FFPE tissues, even from the 2011–2012 collection, did not exhibit a significant reduction in quantity. This underscores the utility of FFPE tissues as an invaluable resource for examining archived specimens and conducting longitudinal studies in CRC research.
The use of two different molecular techniques, including RNA-Seq for FF tissues and nCounter profiling for FFPE tissues, demonstrates the alternative ways of elucidating CRC biology. RNA-Seq allowed for a comprehensive exploration of the transcriptome [17]. nCounter RNA profiling using the PanCancer progression panel provided a targeted approach, facilitating the quantification of cancer-specific RNA transcripts [18]. Furthermore, the nCounter RNA profiling technique is particularly suitable for archival FFPE tissues, which is commonly used in clinical settings and can provide valuable historical data for retrospective studies [18]. These two techniques offer a multifaceted view of CRC biology, spanning the whole transcriptome to specific RNA targets.
In whole transcriptome RNA-Seq analyzes, our comparative results between the two distinct collection periods, 2011–2012 and 2020–2021, provide insights on the potential changes in sequencing reads and gene expression profiles. The consistence observed in sequencing reads, CRC gene signatures, and gene coexpression across two groups of the FF tissues suggests that these specimens remain relatively stable over time. However, further in-depth investigation into pathways uncovers the alterations in ribosome biogenesis in eukaryotes, spliceosome and antifolate resistance across the two groups. Although we report the suitability of approximately 10-year-stored FF tissues from CRC patients for RNA-Seq, these alterations in the pathway associated with the different storage times should be considered extensively.
In nCounter RNA profiling, our comparative examination of FFPE tissues from the two distinct collection timeframes, 2011–2012 and 2020–2021, shows a significant change in gene raw counts, suggesting RNA degradation over time. However, the gene counts become comparable after normalization by the housekeeping genes. Consistency was also observed in the normalized expression of five genes, which were highly expressed in CRC and included in the Nanostring PanCancer panel. Thus, approximately 10-year-stored FFPE tissues were still applicable for nCounter RNA profiling.
Recent studies have underscored the growing recognition of the utility of long-term stored FF and FFPE tissues across diverse tissue types, such as gastric cancer (Type: FF, Storage time: 12 years), human meniscus (Type: FFPE, Storage time: 11 years), breast cancer (Type: FFPE, Storage time: up to 23 years), ovarian cancer (Type: FFPE, Storage time: up to 32 years), and mouse liver (Type: FFPE, Storage time: 21 years [18], [19], [20], [21], [22]. Despite this expanding body of research, there remains a gap in investigations specifically dedicated to CRC and applications in RNA-Seq and nCounter RNA profiling. Thus, our study additionally provides comparative evidence for feasibly utilizing long-term-stored FF and FFPE tissues from CRC patients for RNA-Seq and nCounter RNA profiling, respectively. Our study also suggests two concerns: (i) alterations of ribosome biogenesis, spliceosome, and antifolate resistance pathways in RNA-Seq analysis of long-term-stored FF tissues, and (ii) reduction of raw counts in nCounter RNA profiling of long-term-stored FFPE tissues.
Although this study provides potential insights, it is not without limitations. There are variations in the comparison and utilization of methods across different groups in this study. These differences come from inherent differences in the study populations or specific research objectives for each group. Moreover, methodological considerations, such as sample type/size limitations and technical constraints, have influenced the application of certain methods in some groups. The comparison is done on different specimens, rather than a direct comparison of one tumor material over time. The size and diversity of the patient cohort may be factors to consider for future research to ensure a more comprehensive analysis. Additionally, exploring other sample types, storage methods, durations, and analytical techniques would provide a deeper understanding.
Following are highlights of this study:
First, we report that FF tissues from CRC patients, stored for approximately 10 years, are suitable for whole transcriptome RNA-Seq. However, caution is advised due to observed alterations in pathways associated with ribosome biogenesis, spliceosome, and antifolate resistance.
Second, we report that FFPE CRC tissues stored for approximately 10 years remain suitable for specific RNA profiling using the nCounter pan-cancer progression panel, even though there is a significant reduction in raw counts.
Conclusions
In summary, our comparative results revealed that FF and FFPE tissues derived from CRC patients stored approximately 10.5 years were applicable for RNA-Seq and nCounter profiling, respectively. Importantly, we report two concerns about pathway alteration and raw count reduction in the long-term stored specimens. These findings emphasize the enduring value of biobanks to fuel oncological research.
Funding source: Foundation for Cancer Care, Siriraj Hospital
Award Identifier / Grant number: R016241047
Funding source: Health Systems Research Institute
Award Identifier / Grant number: 63-117
Acknowledgments
This work was highly supported by the Department of Surgery, the Department of Pathology, and the Research Department, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand. The authors thank Ms. Benjarat Thiengtrong and Mr. Panudeth Juengwiwattanakitti for their contributions.
-
Research ethics: Research involving human subjects complied with all relevant national regulations, institutional policies and is in accordance with the tenets of the Helsinki Declaration, and has been approved by the authors’ Institutional Review Board. This study was part of a research series approved by the Siriraj Institutional Review Board (COA No. Si156/2011, Si348/2019 and Si105/2021).
-
Informed consent: Informed consent was obtained from all participants in this study.
-
Author contributions: Pariyada Tanjak: conceptualization. Onchira Acharayothin, Kullanist Thanormjit, and Pariyada Tanjak: methodology. Thanawat Suwatthanarak, Amphun Chaiboonchoe, and Pariyada Tanjak: validation. Thanawat Suwatthanarak, Onchira Acharayothin, Kullanist Thanormjit, Tharathorn Suwatthanarak, Amphun Chaiboonchoe, Apichaya Niyomchan, and Pariyada Tanjak: formal analysis. Thanawat Suwatthanarak, Onchira Acharayothin, Kullanist Thanormjit, Apichaya Niyomchan, and Pariyada Tanjak: investigation. Manop Pithukpakorn, Vitoon Chinswangwatanakul, and Pariyada Tanjak: resource. Thanawat Suwatthanarak, Onchira Acharayothin, Kullanist Thanormjit, and Pariyada Tanjak: data curation. Thanawat Suwatthanarak and Kullanist Thanormjit: visualization. Thanawat Suwatthanarak and Kullanist Thanormjit: writing – original draft preparation. Manop Pithukpakorn, Vitoon Chinswangwatanakul, and Pariyada Tanjak: writing, review and editing. Vitoon Chinswangwatanakul and Pariyada Tanjak: supervision. Vitoon Chinswangwatanakul and Pariyada Tanjak: project administration. Manop Pithukpakorn, Vitoon Chinswangwatanakul, and Pariyada Tanjak: funding acquisition. All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Competing interests: Authors state no potential conflict of interest.
-
Research funding: This research was funded by (i) Health Systems Research Institute (HSRI) of Thailand (number 63–117) and (ii) the Foundation for Cancer Care, Siriraj Hospital (number R016241047).
-
Data availability: The datasets from RNA-Seq and nCounter RNA profiling generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
1. Hossain, MS, Karuniawati, H, Jairoun, AA, Urbi, Z, Ooi, J, John, A, et al.. Colorectal cancer: a review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers 2022;14:1732. https://doi.org/10.3390/cancers14071732.Search in Google Scholar PubMed PubMed Central
2. Coppola, L, Cianflone, A, Grimaldi, AM, Incoronato, M, Bevilacqua, P, Messina, F, et al.. Biobanking in health care: evolution and future directions. J Transl Med 2019;17:172. https://doi.org/10.1186/s12967-019-1922-3.Search in Google Scholar PubMed PubMed Central
3. Annaratone, L, De Palma, G, Bonizzi, G, Sapino, A, Botti, G, Berrino, E, et al.. Basic principles of biobanking: from biological samples to precision medicine for patients. Virchows Arch 2021;479:233–46. https://doi.org/10.1007/s00428-021-03151-0.Search in Google Scholar PubMed PubMed Central
4. Lewis, C, McQuaid, S, Hamilton, PW, Salto-Tellez, M, McArt, D, James, JA. Building a ’Repository of Science’: the importance of integrating biobanks within molecular pathology programmes. Eur J Cancer 2016;67:191–9. https://doi.org/10.1016/j.ejca.2016.08.009.Search in Google Scholar PubMed
5. Alsagaby, SA. Investigating the impact of RNA integrity variation on the transcriptome of human leukemic cells. 3 Biotech 2022;12:160. https://doi.org/10.1007/s13205-022-03223-1.Search in Google Scholar PubMed PubMed Central
6. Malone, ER, Oliva, M, Sabatini, PJB, Stockley, TL, Siu, LL. Molecular profiling for precision cancer therapies. Genome Med 2020;12:8. https://doi.org/10.1186/s13073-019-0703-1.Search in Google Scholar PubMed PubMed Central
7. Kukurba, KR, Montgomery, SB. RNA sequencing and analysis. Cold Spring Harb Protoc 2015;2015:951–69. https://doi.org/10.1101/pdb.top084970.Search in Google Scholar PubMed PubMed Central
8. Goytain, A, Ng, T. NanoString nCounter technology: high-throughput RNA validation. Methods Mol Biol 2020;2079:125–39. https://doi.org/10.1007/978-1-4939-9904-0_10.Search in Google Scholar PubMed
9. Kokkat, TJ, Patel, MS, McGarvey, D, LiVolsi, VA, Baloch, ZW. Archived formalin-fixed paraffin-embedded (FFPE) blocks: a valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobanking 2013;11:101–6. https://doi.org/10.1089/bio.2012.0052.Search in Google Scholar PubMed PubMed Central
10. Hedegaard, J, Thorsen, K, Lund, MK, Hein, AM, Hamilton-Dutoit, SJ, Vang, S, et al.. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One 2014;9:e98187. https://doi.org/10.1371/journal.pone.0098187.Search in Google Scholar PubMed PubMed Central
11. Acharayothin, O, Thiengtrong, B, Juengwiwattanakitti, P, Anekwiang, P, Riansuwan, W, Chinswangwatanakul, V, et al.. Impact of washing processes on RNA quantity and quality in patient-derived colorectal cancer tissues. Biopreserv Biobanking 2023;21:31–7. https://doi.org/10.1089/bio.2021.0134.Search in Google Scholar PubMed
12. Buchauer, L, Itzkovitz, S. cellanneal: a user-friendly deconvolution software for transcriptomics data. J Open Source Softw 2024;9:5610. https://doi.org/10.21105/joss.05610.Search in Google Scholar
13. Hong, M, Tao, S, Zhang, L, Diao, LT, Huang, X, Huang, S, et al.. RNA sequencing: new technologies and applications in cancer research. J Hematol Oncol 2020;13:166. https://doi.org/10.1186/s13045-020-01005-x.Search in Google Scholar PubMed PubMed Central
14. Ghatak, S, Mehrabi, SF, Mehdawi, LM, Satapathy, SR, Sjolander, A. Identification of a novel five-gene signature as a prognostic and diagnostic biomarker in colorectal cancers. Int J Mol Sci 2022;23:793. https://doi.org/10.3390/ijms23020793.Search in Google Scholar PubMed PubMed Central
15. Tanjak, P, Chaiboonchoe, A, Suwatthanarak, T, Acharayothin, O, Thanormjit, K, Chanthercrob, J, et al.. The KRAS-mutant consensus molecular subtype 3 reveals an immunosuppressive tumor microenvironment in colorectal cancer. Cancers 2023;15:1098. https://doi.org/10.3390/cancers15041098.Search in Google Scholar PubMed PubMed Central
16. Ariyannur, PS, Joy, RA, Menon, V, Paulose, RR, Pavithran, K, Vasudevan, DM. Pilot Nanostring PanCancer pathway analysis of colon adenocarcinoma in a tertiary healthcare centre in Kerala, India. Ecancermedicalscience 2021;15:1302. https://doi.org/10.3332/ecancer.2021.1302.Search in Google Scholar PubMed PubMed Central
17. D’Agostino, N, Li, W, Wang, D. High-throughput transcriptomics. Sci Rep 2022;12:20313. https://doi.org/10.1038/s41598-022-23985-1.Search in Google Scholar PubMed PubMed Central
18. Monibi, FA, Pannellini, T, Croen, B, Otero, M, Warren, R, Rodeo, SA. Targeted transcriptomic analyses of RNA isolated from formalin-fixed and paraffin-embedded human menisci. J Orthop Res 2022;40:1104–12. https://doi.org/10.1002/jor.25153.Search in Google Scholar PubMed PubMed Central
19. Zhang, X, Han, QY, Zhao, ZS, Zhang, JG, Zhou, WJ, Lin, A. Biobanking of fresh-frozen gastric cancer tissues: impact of long-term storage and clinicopathological variables on RNA quality. Biopreserv Biobanking 2019;17:58–63. https://doi.org/10.1089/bio.2018.0038.Search in Google Scholar PubMed
20. Pennock, ND, Jindal, S, Horton, W, Sun, D, Narasimhan, J, Carbone, L, et al.. RNA-seq from archival FFPE breast cancer samples: molecular pathway fidelity and novel discovery. BMC Med Genom 2019;12:195. https://doi.org/10.1186/s12920-019-0643-z.Search in Google Scholar PubMed PubMed Central
21. Zhao, Y, Mehta, M, Walton, A, Talsania, K, Levin, Y, Shetty, J, et al.. Robustness of RNA sequencing on older formalin-fixed paraffin-embedded tissue from high-grade ovarian serous adenocarcinomas. PLoS One 2019;14:e0216050. https://doi.org/10.1371/journal.pone.0216050.Search in Google Scholar PubMed PubMed Central
22. Cannizzo, MD, Wood, CE, Hester, SD, Wehmas, LC. Case study: targeted RNA-sequencing of aged formalin-fixed paraffin-embedded samples for understanding chemical mode of action. Toxicol Rep 2022;9:883–94. https://doi.org/10.1016/j.toxrep.2022.04.012.Search in Google Scholar PubMed PubMed Central
Supplementary Material
This article contains supplementary material (https://doi.org/10.1515/labmed-2023-0137).
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Mini Review
- Implementing the ESMO recommendations for the use of circulating tumor DNA (ctDNA) assays in routine clinical application/diagnostics
- Original Articles
- Questionable accuracy of four ELISA kits in serum Netrin-1 measurement
- Evaluation for serum glucose standardization in clinical laboratories of Southern China by consecutive 6 years proficiency testing based on JCTLM-recommended reference methods
- Assessment of long-term stored specimens in the Siriraj Hospital colorectal cancer biobank for RNA sequencing and profiling
- Identification of a high threshold value of serum ferritin in the diagnosis of hemophagocytic lymphohistiocytosis in hospitalized children in China
- Usefulness of neutrophil-to-lymphocyte count ratio, procalcitonin, and interleukin-6 for severity assessment of bacterial sepsis
- Congress Abstracts
- German Congress of Laboratory Medicine: 19th Annual Congress of the DGKL and 6th Symposium of the Biomedical Analytics of the DVTA e. V. together with the 6th German POCT-Symposium
Articles in the same Issue
- Frontmatter
- Mini Review
- Implementing the ESMO recommendations for the use of circulating tumor DNA (ctDNA) assays in routine clinical application/diagnostics
- Original Articles
- Questionable accuracy of four ELISA kits in serum Netrin-1 measurement
- Evaluation for serum glucose standardization in clinical laboratories of Southern China by consecutive 6 years proficiency testing based on JCTLM-recommended reference methods
- Assessment of long-term stored specimens in the Siriraj Hospital colorectal cancer biobank for RNA sequencing and profiling
- Identification of a high threshold value of serum ferritin in the diagnosis of hemophagocytic lymphohistiocytosis in hospitalized children in China
- Usefulness of neutrophil-to-lymphocyte count ratio, procalcitonin, and interleukin-6 for severity assessment of bacterial sepsis
- Congress Abstracts
- German Congress of Laboratory Medicine: 19th Annual Congress of the DGKL and 6th Symposium of the Biomedical Analytics of the DVTA e. V. together with the 6th German POCT-Symposium

