Abstract
Twenty-five years of LOINC (Logical Observation Identifier Names and Codes) and almost 20 years of experience with the implementation of LOINC in Germany – without having so far achieved a binding national definition of or a relevant routine use of LOINC in laboratory data communication. This article sketches the development of LOINC use in Germany since the year 2000 on the basis of grey literature. For the first time, the use of LOINC in Germany is experiencing a significant impetus at the national level: On the one hand, the current health legislation with its stipulations for a legally defined electronic patient record provides the necessary framework for nationwide stipulations; on the other hand, there is a significant impulse from the German Medical Informatics Initiative (MII) out of the medical research field for implementing a uniform LOINC subset. In recognition of the 25th anniversary of the LOINC nomenclature (1995–2019), the article traces the emergence of LOINC – which is characterized by interactions between European (EUCLIDES, READ, NPU) and US (HL7, LOINC, SNOMED CT) developments and the interplay of various standardization initiatives. Different national definitions and e-health strategies resulting from this history will be a challenge for the future e-health harmonization in the EU. The concerns of medical research and biobanking must be taken into account here, since the standardization of lab data according to international nomenclatures is of utmost importance for them.
Reviewed Publication:
Kiehntopf M. Edited by:
Introduction
LOINC (Logical Observation Identifier Names and Codes) is an international nomenclature for medical observations and measurements. Its objective is a language-, system- and institution-independent, universally unambiguous identification of medical analyses and descriptive content, which enables computer-aided automated data processing. Originally, LOINC was developed for the identification of laboratory parameters. Although the use of LOINC now extends far beyond this area, LOINC has its special strength in this domain. The catalog of LOINC codes has been publicly available and in the public domain [1] since 1995; it is compiled and maintained by the LOINC Committee at the Regenstrief Institute, a private non-profit research organization at the Indiana University School of Medicine (IUSM) in Indianapolis, United States, supported by representatives from research, industry and the US government [2].
How LOINC came to be
The origins of the development of LOINC date back to the year 1983: In this early phase of computer use in medicine, C. McDonald, later the “father of LOINC”, and colleagues at the Regenstrief Institute of the Indiana University School of Medicine pointed out in an editorial to the American Medical Association (AMA) that – in addition to other identifications, for example for exchange formats and patient identifiers – there is a necessity for a set of unequivocal identifiers for clinical observations to make a future automated data exchange between medical computer systems possible. McDonald drew a parallel back then to grocers, who introduced universal food product codes nationwide in the 1970s, paving the way for electronic cash register and merchandise management systems in food retailing. Similarly, in the midst of the era of the first, still purely mainframe-based patient record systems, McDonald saw the perspective for medicine – falling hardware and software costs would soon lead to the situation that every physician’s practice would be able to afford such a system, and thus clinical data interchange (CDI) would become a necessity [3]. The fact that his editorial was rejected nine times between 1981 and 1983 [4] before it was published shows that this view was hardly undisputed – the editors provided an elucidating note: “Perhaps this article will provoke the discussions that will result in appropriate actions” [3]. In fact, this impetus resulted in the first work on a corresponding standard [5], which was done in 1984 under the umbrella of the American Society for Testing and Materials (ASTM) in a Subcommittee E31.11, “Standards for the Exchange of Clinical Data”, established for this purpose.
Early on, for pragmatic reasons, they focused on clinical laboratory results data: These are not only of high clinical relevance, but are also available in a precisely defined, structured form. In 1988, the standard developed by the ASTM was adopted and published as E1238-88 “Standard Specification for Transferring Clinical Laboratory Data Messages Between Independent Computer Systems” [6] – as the first standard for clinical data which has undergone a public balloting and consensus process, in the way it complied with the ASTM policies at that time and thus with the criteria of the American National Standards Institute (ANSI), and in the way HL7 still practices it today in a similar form [7]. ASTM E1238 was incorporated as Chapter 7 in the standard HL7 V. 2.1, the further development of the first version of HL7 published in 1987. In the following years, further work on the standard for laboratory data communication shifted increasingly to HL7; the history of this merging is extensively presented elsewhere [8].
ASTM E1238 successfully addressed the syntax of standardized messages – in the form of the logic of the OBR and OBX segments adopted by HL7 for mapping order and observation – and the thus implicitly associated information model. The semantics were left open for many data elements. For the identification of laboratory testing, the ASTM E1238 standard proposed a relatively simple expansion of the current procedural terminology (CPT codes), which have been published since 1996. However, this approach proved itself to be insufficient, especially since it lacked an unequivocal standardization of other clinical variables [4], [9].
In parallel to these developments in the US, the European Union began promoting the Advanced Informatics in Medicine (AIM) framework program during the second half of the 1980s (1988–1990 AIM I, 1991–1994 AIM II). In the context of the so-called “AIM Exploratory Action”, 42 projects were funded from the beginning of 1989 onwards; which in many aspects developed important foundations for health telematics in Europe [10], [11], [12].
Within this framework, the EUCLIDES project (European Clinical Laboratory Data Exchange Standard) developed a European standard for the exchange of clinical laboratory data between various medical information systems in different European countries and languages. EUCLIDES also dealt, as ASTM E31.11 did, with all questions of data transfer mechanisms and message syntax – as well as with the development of a medical code system for use in these messages [13], [14], [15]. It is interesting that here, in addition to the communication of findings in the context of patient care, further areas of application for standardized laboratory data exchange have already been addressed: external quality assurance, interfaces between analytical instruments and laboratory information systems, and aggregation of data for regional and international evaluation for health system analysis purposes. The EUCLIDES semantics were initially intended to cover the areas of clinical chemistry, toxicology, hematology and immunology as well as microbiology and serology. For the EUCLIDES code system, the project group around G. De Moor developed six different vectors, which made it possible to differentiate among laboratory tests [14], [15]. This anticipated the later multi-axial system of LOINC, as shown in the following comparison in Table 1.
Vectors or axes of the EUCLIDES (1992) and LOINC (1995) nomenclatures in comparison.
| EUCLIDES (1992) | LOINC (1995) |
|---|---|
| Tests (No. 1): analytes/procedures/function tests/ratios | COMPONENT (No. 1): component/analyte |
| Specimens (No. 2): types/locations/collection procedures | SYSTEM (No. 4): system type/sample type |
| Lab Procedures (No. 3): basic analytical methods/reagents/temperature/equipment | METHOD TYPE (No. 6) |
| Units (No. 4): denominators/numerators | – |
| – | SCALE TYPE (No. 5) |
| Kinds of Quantities (No. 5) | PROPERTY (No. 2): kind of property / kind of quantity |
| Coded comments (No. 6) | – |
| – | TIME ASPECT (No. 3) |
With the 1994 version 4.0 of the EUCLIDES OpenLabs Coding System, >6500 codes and 40 object classes (ANALYTE, SAMPLE TYPE, METHOD, REAGENT, PROPERTY, UNIT, and many others) were distinguished for the laboratory especially clinical pathology area. Each object class had its own code. One lab measurement was thus expressed by several codes [15]. An example:
| Haemoglobin: | 02410 | Haemoglobin | = object class ANALYTE |
| 01256 | kg/L | = object class UNITS | |
| 01062 | Mass Concentration | = object class PROPERTY | |
| etc. |
Its own foundation was established to further develop EUCLIDES (EUCLIDES Foundation International, Belgium), and the syntactic approaches of EUCLIDES flowed into the work being done at that time by the Technical Committee for Health Informatics (TC 251) of the European Committee for Standardization (Comité Européen de Normalisation, CEN) on an EDIFACT-based European messaging format for healthcare system. Early on, this led to contact and cooperation between EUCLIDES and ASTM E31.11 in the US.
An analysis by the Board of Directors of the American Medical Informatics Association (AMIA) in 1994 represented an important basis for further work in the US; this identified the areas of application for which suitable code systems were already available in the context of electronic communications among hospitals, medical practices and diagnostic laboratories, as well as in the development of electronic medical records. For diagnostic procedures and examinations, the analysis came to the conclusion that there was still a lack of suitable identifiers which could be used in particular in the newly established HL7 syntax at that time; it was urgently necessary to define these centrally [16]. The EUCLIDES approach was seen as important groundwork, but too complex for further work: According to the EUCLIDES terminology, each form of expression in the vectors was represented by a corresponding digit in the code, which allowed a great power and variety of concepts to be identified by codes. At the same time, however, there was no sufficiently large list of easy-to-use, medically relevant and verified pre-coordinated codes available. This led to the development of the LOINC catalog in 1995 [17]: The various axes remained, but the logic of code generation was simplified and, in particular, emphasis was placed on providing as comprehensive and unambiguous a list of diagnostic observations and measurements of practical relevance as was possible – including beyond the laboratory sector. A first version of this new catalog was published in 1995 [2].
Since 1995, LOINC has proved itself in many systems and hospitals in the US, and also internationally, especially for the clear identification of laboratory and vital parameter determinations. More areas of application were soon being addressed with the growing LOINC catalog, such as, for example, clinical observations and medical documentation, electrophysiology and radiological and microbiological examinations [18].
Further development of LOINC into the “lingua franca” of clinical data exchange
The Regenstrief Institute makes the LOINC catalog available for download on a separate website as an access database as well as in other file formats (TXT or CSV, PDF) [1]. Every year, one or two update versions are published, which incorporate new codes and examinations and, to a lesser extent, obsolete entries are also removed or supplemented. In this process, the scope of the catalog has increased considerably over the course of the past almost 25 years: While version 1.0 was published in April 1995 still with 5900 entries, the database version 2.6 in June 2019 included around 91,388 entities. Figure 1 illustrates the evolution of the database (according to [19]).

Development of the LOINC database 1995–2019.
In addition to the database and the LOINC manual, which provides the definitions and application instructions, the Regenstrief Institute provides a variety of other auxiliary tables and tools all around the use of LOINC, including frequently asked questions (FAQs), training materials (slides), online training and testing routines. In particular, the software tool RELMA (REgenstrief LOINC Mapping Assistant) helps to find the appropriate identifier for a local examination in the very extensive LOINC database and to save the assignments. It also offers additional logic and hierarchical drill-down mechanisms. RELMA thus offers support for mapping to LOINC that goes beyond the native database.
The codes as well as the tools are copyright protected, but may be used freely if you adhere to the prescribed rules.
In principle, LOINC encodes only the semantics of a clinical examination or observation; in many cases this can be expressed by a single LOINC code. Accordingly, LOINC can be used with virtually any communication protocol and any messaging and document standard [27], [28]:
HL7 V2 (OBX Segment Field OBX-3.1=Observation Identifier)
HL7 CDA
HL7 FHIR
LDT (field 8410=“Test-Ident”) – communication protocol used in the sector of German statutory health insurance (SHI) accredited physicians –
CDISC LAB, with restrictions also CDISC SDTM
CCR
open EHR
Integrating the Healthcare Enterprise (IHE) profiles
It is important that an identification can be transmitted that the code used for the investigation is a LOINC code. To the extent that the respective protocol allows, the LOINC code can also be transmitted in addition to a readable proprietary display name that corresponds to the respective domestic usage.
Accordingly, LOINC has been recommended for use early on, since the 1990s, by professional associations in the US and international standardization initiatives; thus, for example, HL7 and the IHE recommend LOINC as the preferred code system at least for laboratory data. Since 2005, CDISC has also referred (as part of the cooperation begun with HL7) to the potential for using LOINC codes within the CDISC formats; the Food and Drug Administration (FDA) is considering recommending the use of LOINC submission of drug approval studies. Finally, since 2012, there has been formalized cooperation on an international level between the International Health Terminology Standards Development Organization (IHTSDO, also known as SNOMED International), which publishes SNOMED CT under license, and the Regenstrief Institute, with the declared objective of establishing a uniform order entry and result reporting with both code systems and of coordinating the further development of semantic standardization. In this process, it has been made clear, firstly, that LOINC is not merging with SNOMED CT; secondly, that all rights to LOINC remain with the Regenstrief Institute and that the use of LOINC will not be affected by the license obligations for SNOMED CT [20].
During the past 25 years, the use of LOINC in various application areas has become increasingly accepted internationally. The Regenstrief Institute states on the LOINC website that worldwide there are around 85,000 LOINC users in 176 countries, whereby LOINC has become the “lingua franca of clinical data exchange” [1].
Construction and structure of LOINC
The construction and structure of LOINC are explained extensively elsewhere [19], [21], [22], [23]. Only the most important characteristics are presented here in an overview.
According to the LOINC concept, any laboratory test, clinical observation or medical examination is unambiguously described by means of six axes: COMPONENT, PROPERTY, TIME ASPECT, SYSTEM, SCALE TYPE and METHOD TYPE. These six axes are represented in corresponding columns of the LOINC database (see Figure 2, according to [24]).

The six axes of LOINC nomenclature (below: the names of the LOINC database columns).
With regard to the sixth axis, the method of examination, it is restrictive that this is only to be indicated as far as it is necessary for the unambiguous determination of the measurement or examination in question and this distinction has fundamental clinical relevance. This column is therefore often empty.
Together, these six axes yield, according to the LOINC nomenclature rule, the unique full generic identifier for the examination (“fully specified name”).
Each type of examination, measurement or observation, that differs in any of these six axes, receives its own identifier, the LOINC code [database field LOINC_NUM; hereinafter also referred to as LOINC-ID if used as an identifier in data transmission and data pooling], and its own entry in the LOINC database. The LOINC codes are assigned consecutively for the entries; the codes do not reflect relationships between concepts, unlike, for example, in a classification. It consists of a numeric multi-digit code with a trailing check digit, which is formed according to a Mod10 algorithm. The procedure is described in the Regenstrief Institute’s LOINC User Manual and by the LOINC Committee [21].
In summary, each clinical observation or medical examination is thus represented by seven elements: the preceding LOINC code and the formal name consisting of the six axes described above (“fully specified name”). Figure 3 illustrates this using the example of (semi)quantitative measurement of urine sugar by means of test strips (according to [19]).

LOINC name and LOINC code.
By means of the six axes, it is possible to distinguish among determinations of the same substance, for example glucose, in different testing materials, such as in serum, urine or cerebrospinal fluid, and also by means of different methods, on various scale types and in various properties. Conversely, it is necessary to know and define all six axes in order to clearly arrive at the correct unique LOINC code for the examination in question. In the PROPERTY axis, only the property is clearly defined – not the dedicated unit of measurement or its notation. If, for example, you consider a glucose measurement, it can thus be shown that the determination of a molar substance concentration [PROPERTY=SCNC] will be distinguished from a mass concentration [PROPERTY=MCNC]; both determinations thus receive different LOINC codes in the same sample type. For the latter, however, it is irrelevant to the uniqueness of the LOINC code whether this is given in the unit g/L or in mg/dL, and how these units are expressed in machine-readable form.
To obtain internationally unambiguous machine-readable coding of units of measurement in order to transmit findings, it is recommended to likewise use the standard unified code for units of measure (UCUM), also issued by the Regenstrief Institute [25], [26].
Lists of common UCUM terms are delivered along with the LOINC accessory files, and there are tools to help you assign the correct UCUM unit to a given PROPERTY and vice versa when using LOINC. However, the unit of measure is not a defining component of the LOINC code.
In addition to the cited seven defining components, there are many more fields for each entry in the database, which vary considerably in number over the versions of the database; however, these do not contain any further definition criteria, but rather merely facilitate dealing with and doing research in the database. These include fields with alternative identifiers and functional synonyms, fields for arranging the catalog into specific classes (e.g. “laboratory” vs. “clinical”), fields with supplementary linked information on a substance or determination (e.g. species information, chemical formulas, and in previous editions of the database also molecular weights, and mappings to other code systems such as SNOMED CT or EUCLIDES), as well as fields that serve the traceable version management of the LOINC database. As a general rule, LOINC codes are never reassigned or deleted; rather they are left in the database for reasons of making the different versions traceable, but put into a non-active or non-recommended status. For fields that are no longer active or no longer recommended, there is moreover a recommended mapping in the database to the corresponding new LOINC codes. If the required entry is missing for a to-be-coded examination, then the inclusion of a new code in a regulated procedure can be requested from the LOINC Committee. In this process, the new inclusions do not follow theoretical considerations, but the practical needs of the international community.
Challenges and improvements
A significant challenge in the use of LOINC lies in the enormous scope and power of the catalog: It makes possible a highly granular description and unequivocal identification of examinations, observations and measurements in what has meanwhile become more than 90,000 concepts. The scope of application extends not only to laboratory parameters but also to vital signs and other clinical properties. The breadth of the vocabulary achieved is impressive – almost every variant for every examination or observation can be found in the catalog. The breadth of being able to map almost everything is an advantage on the one hand, but at the same time also a complication: When determining which LOINC code represents an examination which is to be documented, it requires a precise selection from the existing vocabulary. The “mapping” of the local examinations and measurements to the LOINC nomenclature therefore becomes a time-consuming step requiring both medical domain knowledge and an equal knowledge of the LOINC logic. The following example illustrates just how great the mapping-related “agony of choice” in the LOINC database is, due to the very large set of pre-coordinated concepts and codes: For all practical variants of glucose determinations alone, the LOINC database offers around 140 entries – and thus alternatives to choose from. This does not even take account of the glucose tolerance tests (CLASS=CHAL) – for this purpose there are another some 560 codes available – or the many determinations of the glucose-converting enzymes. For different glucose determinations in serum or plasma alone, about 20 codes are available [19].
Accordingly, the LOINC Committee has in recent years gradually expanded the potential ways of supporting mapping and reducing mapping efforts, including by the constant development of the RELMA tool and the introduction of mapping to “display names” and “consumer names”, as well as by showing references and hierarchies within the catalog. These possibilities have been expanded by the introduction of “LOINC parts” in recent years.
For this purpose, a single part of a LOINC name, a dimension of the “fully specified name”, is considered a “LOINC part” and represented by an “LP code”. For example, glucose – a COMPONENT, a dimension of many LOINC terms – has the LP code LP14635-4. Thus it is possible to represent, independently of synonyms and natural languages, the common feature in many LOINC entries, that glucose is to be measured. The same can be done for the other dimensions or axes of the relevant LOINC terms. Accordingly, you can also re-assemble them from the LP codes. Thus, for example, for the glucose determinations in serum, emerges as mass concentration (LOINC code 2345-7) or as molar substance concentration (LOINC code 14749-6):
LP31388-9.LP31399-6.LP14635-4.LP42107-0,7,LP42107-0, 2345-7, Glucose SerPl-mCnc
LP31388-9.LP31399-6.LP14635-4.LP42107-0,9,LP42107-0,14749-6, Glucose SerPl-sCnc
The “LOINC parts” are used in the maintenance of the LOINC catalog for algorithmic generation of names, for synonym management and for translation, and also for mapping multi-axial hierarchies. These are managed outside the LOINC catalog in their own tables. In particular, the RELMA tool database uses these and other options to map hierarchies and references.
One of the most recent developments is the concept of the “LOINC groups”, introduced in 2017, which is of particular interest for all questions concerning data aggregation and the evaluation of aggregated data sets. The core idea is to use the “LOINC Groups”, which are available either as LOINC Groups File in table format or as FHIR ValueSets, to combine similar LOINC entries in a cascaded approach (“ParentGroups” – “Groups”) for generic questions and to express them with uniform identifiers in order to exchange such groupings in a machine-readable and interoperable way. Both for order entry processes as well as for research evaluations in particular, a certain lack of detail may be necessary to be able to aggregate data without having to go to the granularity level of each LOINC code. For example, it may be irrelevant for evaluations of the number of glucose determinations performed in blood whether they were performed in specified venous blood or in unspecified blood, serum or plasma (all of which have their own LOINC code). Such an analysis can draw on a now standardized “LOINC group”, which can be unambiguously expressed by a “Group ID”. In Version 2.66 of 2019, there are already around 6500 such “LOINC groups” (with about 25,500 terms included) available. However, a reality check of the usefulness and manageability of this mechanism in medical research projects is still pending.
For non-English-speaking countries, there is still the obstacle of translating the LOINC catalog into the respective national language. RELMA can use translations of the LOINC terms, to the extent these are available. This enables the user who is searching for medical terms in his own language to find quickly and unambiguously the correct LOINC term for the examination or observation which he would like to map in his documentation or data communication.
Distinction from NPU, SNOMED CT and CDISC
Nothing is more problematic for standardization than a variety of rival offerings from different standardization organizations: Freedom of choice and uncertainty lead to variance or even to waiving the introduction of standards. This can only be resolved by binding definitions and governance. The old sentence attributed to different authors applies: “The problem with standards is that‘s’at the end”.
Such double offerings as well as overlap between different terminologies also exist in the field of laboratory data. In particular, NPU and SNOMED-CT are to be mentioned here, as well as partially CDISC, Standard PREanalytical Code (SPREC) (see below), DICOM and coding systems and billing code numbers for procedures. Only worth mentioning for historical reasons are the semantic data standards VITAL (Vital Signs Information Representation, CEN ENV 13734/35, today a component of ISO IEEE 11073) in the field of electrophysiology [29], Read Codes (published in the UK in 1986 and today replaced by SNOMED CT in the NHS sector) [30] as well as Names-Lab©, a French development parallel to LOINC and NPU, which in particular addressed the laboratory order entry process, but did not become established [31].
SNOMED CT has already been gone into. In principle, almost every pre-coordinated term in the “big hawker’s tray”-like LOINC catalog can also be expressed post-coordinated as a SNOMED CT term. In addition to the licensing framework conditions, it remains open whether the usage of the distinctly more powerful but also more complex SNOMED CT tool is in all cases the correct answer to the fact that already the complexity of mapping to the much simpler LOINC nomenclature has so far been one of the main obstacles to further dissemination. For some cases of application, however, the use of SNOMED CT instead of LOINC or a combination of both in fact may be advantageous: Thus, there are increasing numbers of project experiences and recommendations that in the microbiological analysis of different pathogens, the latter should not be represented by individual LOINC codes, but rather the pathogen should be understood as the result content and expressed using SNOMED CT terms. Similarly, SNOMED CT terms can be used more meaningfully to express anatomical details of a sample [32]. Both ways are certainly transferable and interoperable, given the international cooperation already mentioned [20]. It will be important that, with the desired convergence, fundamental changes in standards and thus problems with backward compatibility be avoided in order to create confidence in the future-proofing and manageability of these standards.
In contrast, NPU (Nomenclature for Properties and Units) is a development completely parallel to LOINC: NPU goes back to the first considerations of Dybkaer and Jørgensen for the International Federation of Clinical Chemistry and Laboratory Medicine (IFCC) and the International Union of Pure and Applied Chemistry (IUPAC) of 1966, on to how quantities and units in clinical chemistry could in principle be expressed in an internationally uniform way [33], [34]. On the basis of these repeatedly updated recommendations, IUPAC and IFCC on their part published an international standardization recommendation for syntactic and semantic specifications for measurements and units in clinical laboratory medicine in 1995 [35] and developed it further in a series of technical papers and recommendations during the following years into the Nomenclature for Properties and Units (NPU) [36], [37], [38], [39], [40], [41], [42], [43]. In 1997, this term was used for the first time and NPU codes were spoken of, derived from the name of the responsible joint IFCC and IUPAC boards, the Committee on Nomenclature, Properties and Units (C-NPU) [36]. In contrast to LOINC (and for example, also to the Read codes and EUCLIDES), computer-based data exchange was not the initial focus. Around the year 2000, however, this standardization approach of international chemical associations was also taken up in international health telematics.
NPU is a multi-axial catalog for laboratory determinations (but does not go beyond this) and therefore shows similarities to LOINC (for comparison see Table 2). Here again, various dimensions related to the LOINC approach were summarized in one code:
System (examined material)
Component (analyte)
Type of property
Measurement result (number of digits of a numerical value and unit)
Comments, if applicable
Comparison of the axes of the LOINC (1995) and NPU (1995 ff.) nomenclatures.
| LOINC (1995) | NPU (1995 ff.) |
|---|---|
| COMPONENT (No. 1): component/analyte | COMPONENT (No. 2): part of a system |
| PROPERTY (No. 2): kind of property/kind of quantity | KIND-OF-PROPERTY (No. 3) [+ KIND-OF-QUANTITY: unit, procedure] |
| TIME ASPECT (No. 3) | – |
| SYSTEM (No. 4): system type/sample type | SYSTEM (No. 1) |
| SCALE TYPE (No. 5) | – |
| METHOD TYPE (No. 6) | – |
All dimensions can rule-based be extended by specifications and thus be hierarchized. Within the dimensions, the general rules of the IUPAC nomenclatures are strictly applied. The NPU code expresses the information on the system, component, property and unit in a number – five digits which are identified with the prefixed letters “NPU” [43], [44], [45], [46].
Example of a NPU code: glucose in serum:
NPU02187 B – Glucose; subst. c.=? mmol/L
NPU code+short definition
NPU is also made available as a database or table-based code list in the public domain. The catalog also displays a versioning of the codes that go back to the first release in 1996. The current database contains 27,226 (valid and historical) codes. Translations from English into some other languages are available. German experts were involved in the development of NPU [46], but so far there is no German translation.
NPU was developed, as described above, within an international framework, but it was strongly influenced by European-Scandinavian input from the start. As a consequence, Denmark and Sweden have early adopted NPU as part of their national e-health strategies. The Danish health authorities are also responsible for the international publication of the NPU database and maintain a central website on NPU terminology [47].
NPU therefore is a serious alternative candidate to LOINC, which, like LOINC, is already being considered in some countries for their respective national e-health strategies. In order to avoid uncertainty and divergent developments, in 2009 IHTSDO and the Regenstrief Institute endeavored to bring about cooperation and convergence with C-NPU [48], [49]. Irrespective of the willingness to cooperate, there are two different catalogs, and each project, as well as the entities responsible for national specifications, must consider which catalog to use.
In the German-speaking countries, the first such assessment was made at the start of the previous decade by the Swiss CUMUL project (see below) of the Centre Suisse de Contrôle de Qualité, which decided on LOINC for pragmatic reasons: NPU is held to indeed be more coherently defined, but at the same time, in practice it effectively excludes all usual tests which do not follow the IUPAC rules in nomenclature and units. Furthermore, all clinical observations outside the laboratory chemistry, to which NPU is by definition limited, are excluded. The greater flexibility in perspective to obtain codes for missing terms, the greater scope of the database and the perspective for transatlantic standardization all spoke for LOINC at the time [51]. Sixteen years later, a similar assessment is also the basis for the definitions of the German Medical Informatics Initiative (MII) (see below): Bietenbeck et al. compare in detail the representation of laboratory data when using LOINC, NPU and SNOMED, and additionally emphasize that NPU supports the most consistent metrology and expresses both the analysis as well as the result value, whereas LOINC merely represents the analysis and allows a greater degree of freedom in terms of the units – for which reason additional definitions, for example on UCUM, are required. Both approaches, in contrast to SNOMED CT, do not provide any relations between their codes, which is something that LOINC cures with its “LOINC groups” approach. Overall, LOINC offers the easiest manageability [52]. Therefore, in the European region it will therefore remain an important task to consolidate the different national preferences for NPU (e.g. Denmark) or LOINCs (e.g. Switzerland [53], Austria [54], long before Germany) in an interoperable way and to provide manageable services for mutual transfer, in order to ensure cross-border data traffic in the EU on this point.
There have also been obstacles in the area of clinical research for a long time, due to overlap of standards: The family of CDISC standards, which is particularly widespread in the pharmaceutical industry and among regulatory authorities, defined its own CDISC Controlled Terminology in its specifications, and the most common CDISC data format relevant for clinical trial submissions in the field of clinical research, CDISC Study Data Tabulation Model (SDTM) Data Exchange & Tabulation Standard, initially did not provide for the use of LOINC. In addition, structures of the CDISC SDTM fields – unlike those of CDISC LAB – were defined in such a way that they, along with LB.LOINC, the data element of LB domain introduced in the meantime, do not allow a completely disjoint mapping of LOINC axes [55]. In 2015, the FDA announced in the US that it would not only accept LOINC in the submission data of clinical trials, but also would call for the use of LOINC [56]. In 2017, a joint LOINC working group of the FDA, NIH, CDISC and Regenstrief Institute published a joint recommendation on how to map LOINC to the various CDISC SDTM variables and developed mapping guidelines based on the subset LOINC 2000+ [57]. Starting from March 2020, the FDA will be considering LOINC as the expected format for laboratory data from clinical trials. A further convergence and a greater significance for LOINC in the field of drug approval and regulatory affairs is therefore also to be expected internationally.
The benefits of LOINC: data integration and data pooling
The benefits of LOINC standardization are particularly evident where clinical data (laboratory determinations, vital signs, clinical assessments and other test results) from different sources (submitting laboratories, subsystems, direct data transfer by medical examination equipment, manual entry into the relevant database, etc.) are to be merged and therefore should be consolidated (see Figure 4, according to [27], [28]).

Data integration and data pooling: LOINC replaces the proprietary identifiers for laboratory measurements from various data sources which as shown in the Figure, and thus makes it possible to consolidate the data – whether for an electronic patient record, for a study database, a research data warehouse or any other integration platform.
The use cases are manifold:
hospital information systems and medical practice systems in communication with external diagnostics;
PDMS systems in anesthesia and other clinical documentation systems with broad direct connection of measuring devices;
telemonitoring control centers that receive data from various sensors and diagnostic systems;
the patient’s personal health records which are intended to take over data from various service providers;
study clinical trial databases and other research databases that consolidate data in multi-center research projects;
data consolidation as part of quality assurance;
data consolidation in the context of public health and health reporting.
In all the scenarios listed as examples, the receiving and data-converging IT systems benefit when LOINC-standardized clinical data is to be transmitted, thus making it possible to avoid manual effort and assignment errors. The prerequisite is, of course, that the sending systems are able to transmit (correctly mapped) LOINC codes along with their data results.
For laboratory medicine, and also for intensive care medicine – both of which use a high rate of instrument-based diagnostics, each time with different data sources – there are highly relevant scenarios that would benefit from the use of LOINC:
electronic transmission of laboratory results data (observation reporting), for example in the communication between the laboratory information system and the hospital information system;
laboratory requests as part of a fully electronic order entry process (where the request often requires a lower level of granularity of the coded information than the transmission of findings – which is why the multiaxial hierarchy and LP codes in the LOINC system are particularly important for this purpose);
data pooling in inter-laboratory comparison tests within the framework of quality assurance in laboratory medicine;
accounting and billing of diagnostic services – which requires appropriate mapping to accounting systems and billing code systems;
export of data for medical research purposes (clinical research, health services research);
as well as the prospective export of data to the patient or the citizen himself – to support automated transfer into layman-friendly texts via a LOINC standardization.
Implementation of LOINC in Germany
The first German-language application of LOINC was made in Switzerland: As part of the CUMUL project of the European Laboratory Medicine (ELM), a first preparatory work in Europe in this field which started in 1997, LOINC codes were selected, annotated and trilingual translated – including into the German language – for purposes of laboratory quality assurance in the Quality Control Center Switzerland (CSCQ). The intention was to establish permanent “reportable names” for the selected LOINC terms, all of which are from the “Laboratory LOINC” area, as identifiers in four languages (English, French, German, Italian) [50], [51], [58].
In 2000, the HL7 user group made LOINC known to the broader professional public in Germany in its communications for the first time [59].
German standardization committees (DIN NAMed FB G 1 & G 3, HL7 user group) subsequently made references to LOINC and in 2001 came the first industrial implementations in projects and products as well as the first use of LOINC in routine clinical operations, first in laboratory medicine in the Westphalia State Insurance Institution [24], [27], [60] and in 2002 at the Kiel University Hospital, where a complete LOINC mapping of all parameters of the central laboratory, including transmission of LOINC-standardized data to the subsystems at the clinical workplace, was done for the first time in a routine operation at a university hospital in Gemarny [28], [61]. Parallel to this, the evaluation of LOINC began in a clinical pharmacology treatment-monitoring project at the Erlangen University Hospital. Somewhat later followed the usage of LOINC in research and care in anesthesia and intensive care at the Jena and Gießen locations [62]. Some groups of laboratory physicians and the German National Association of Statutory Health Insurance Physicians (KBV) sporadically tested with LOINC in order to review its use in the general practice area. In 2003, the software product manufacturers also began consultations on this subject within the Association of Manufacturers of IT Solutions for the Healthcare Sector (VHitG) (today Federal Association of Health IT [BVITG]).
Starting from 2004, industrial products with LOINC implementations were presented at the annual industrial fair Information Technology Design (ITeG) (later Connecting Healthcare IT [conhIT] and today Digital Medical Expertise & Applications [DMEA]).
In early 2004, an informal “LOINC User Group” was brought into being, where there was an exchange of experiences among users from the various domains [63]. In 2005 followed the founding of the “Standardized Terminology in Medicine (STM)” project group within the German Society for Medical Informatics, Biometry and Epidemiology (GMDS); at the end of 2006, the Technical Committee (TC) Terminologies of the HL7 User Group Germany was founded (later part of the Joint Interoperability Forum of HL7 and IHE Germany). All groups contributed to the exchange of experiences, which initially focused on the use of LOINC to transmit findings in the clinic laboratory [64], [65], [66] and only considered order entry processes somewhat later [67]. A first migration of a laboratory system was implemented in 2006 while retaining the LOINC-standardized interface with the clinical workstation system – almost without the users at the clinical workstation noticing – because the transmitted data retained the same semantics [65], [66]. Considerations of using LOINC for a cross-border standardized schedule of services for billing purposes [64], however, were not pursued. The German Federal Agency for Medical Documentation and Information (DIMDI) was motivated to also look into LOINC within the framework of classification systems not stipulated by law [68]. Since 2005, DIMDI has been hosting the LOINC catalog for Germany, providing some basic information and acting as a contact point for reporting missing codes to the Regenstrief Institute [69]. In 2005, the LOINC User Guide was translated into German and made available on behalf of DIMDI [70]. In 2007 and 2009–2010, DIMDI carried out the translations of part of the LOINC terms into German, using preparatory work by the Vienna Hospital Association from Austria and from the afore mentioned projects in Germany, and also from the Swiss CUMUL project. The translations were subjected to both terminological and medical quality assurance. The translated LOINC terms are today part of the RELMA database. However, these are only available for a smaller part of the LOINC database and have not been maintained up to the present. Also, despite existing obstacles to the use of LOINC, competencies for using the RELMA tool were developed comparatively late due to the mapping effort involved [71].
In the field of medical research, notice of LOINC was initially delayed, especially in the area of clinical trials, where other standards and terminologies were in the foreground (e.g. the CDISC standard family, MedDRA, ATC; and additionally SPREC in the biobanking area). In January 2007, an expert workshop, initiated and organized by the Technologie- und Methodenplattform für die vernetzte medizinische Forschung e.V. (TMF) in cooperation with GMDS and HL7, initiated expert discussion on the prospects of using the terminology in medical research and what strategic definitions would be necessary for this [72]. The potential of LOINC for research was explored for the first time; with particular interest in the extent to which LOINC can be used within the CDISC standards by both the pharmaceutical industry and the academic community, as part of a future “common language” for research and health care [73], [74]. The German-speaking CDISC user group also started to address this issue in their meetings in 2009. Likewise, a metadata repository (MDR) jointly developed by several academic partners under the umbrella of the TMF provided for the possibility of referencing LOINC, which in principle would allow the reuse of already LOINC-standardized data elements for research projects.
Despite all these initiatives by experts, the routine use of LOINC in Germany has hardly progressed for several years. The degree of use barely increased between 2003 and 2013, although already early on there had been no lack of corresponding recommendations in strategy papers on health telematics and e-health in Germany. From the Telematics Expertise of the German Industry 2003 [75] on the first state-mandated “bit4-health” concepts to introduce an electronic health card starting from 2004 [76] up to the E-health Planning Study on Interoperability of 2014 commissioned by the German Federal Ministry of Health (BMG) [77] – LOINC has always been mentioned as the most important element for creating semantic interoperability and its (mandatory) implementation for specific documentation areas has been demanded. The fundamental importance of a controlled vocabulary for the benefit of a telematics infrastructure in health care was pointed out at an early stage [78]. A TMF expert study commissioned by the German Federal Ministry of Health (BMG) on terminologies in the German-speaking countries offered the analysis, in the TMF’s resulting recommendations in 2015, that in Germany, due to a lack of commitment or incentives for remuneration, there had been no successful dissemination of an international standard for routine operations in Germany, except by the International Classification of Diseases (ICD). Despite its free availability and relatively good controllability, LOINC is developing slowly in the market. Against the background of European Commission initiatives such as Smart Open Services for European Patients (epSOS), in which LOINC codes for the value set were also provided for any cross-border treatment-related data exchange, trans-national cooperation among Germany, Austria and Switzerland was recommended [79]. Austria and Switzerland have implemented these European impulses more quickly and declared that the transmission of LOINC-coded laboratory data is obligatory for some sectors of their national EHR projects (ELGA or ePatient dossiers) [53], [54]. For Germany, on the other hand, it was necessary to put it on record in the most recent recommendations for handling big data in the health sector, commissioned by the German Federal Ministry of Health (BMG) that, due to the lack of binding provisions at the national level and due to the lack of an e-health strategy including aspects of medical research, there is no change to the fact that the use of international semantic standards (such as LOINC and SNOMED CT) continues to make little progress, and that the use of existing data in the health sector is thus susceptible to being hindered [80].
An inventory at the end of the last decade has shown little routine use, but some experience and a great deal of potential for using LOINC in a variety of applications [81]:
laboratory medicine;
intensive care;
medication safety (Arzneimitteltherapiesicherheit [AMTS]);
physician’s discharge letters and medical documentation;
scores and assessments;
clinical and epidemiological research.
To these was added the first experiences in the field of microbiology and health reporting (electronic pursuant to the Infection Protection Act [82]).
Overall, LOINC use in Germany has lagged far behind that in the US over a number of years. Only in recent years has there been any intensification of LOINC usage. At the same time, it is interesting that “Laboratory LOINC” is not the focus of interest, but rather the concepts in “Clinical LOINC” for the simple coding of medical documents and segments in medical documentation, also for categorizing documents in a hospital information system [83].
The more CDA documents with structured medical content are used, the more frequently LOINC codes and the LOINC clinical document ontology will also be used in German projects and specifications. With the increasing use of HL7 CDA and, in the past 3 years, FHIR, Laboratory LOINC codes have also found increasing usage, for example in protocols for emergency medicine [84].
By means of current legislation in Germany (E-Health Act [EHG], 2015; Appointment Service and Health Care Act [TSVG] 2019; Digital Health Care Act [DVG] 2019) not only has a comprehensive electronic patient record been introduced for all insured persons in Germany, but also the National Association of Statutory Health Insurance Physicians (KBV) and the Gematik, the German Agency for Telematics Infrastructure in Health Care, have been given the authority to determine how to define the contents of the electronic patient record and to establish standards. It is to be expected that this will result in a significant boost in the coming years, which will further advance laboratory data standardization in Germany using LOINC.
Impulse from research: the German Medical Informatics Initiative (MII)
The most important impulse for using LOINC in Germany is currently coming from medical research. This approach is special to the extent that developments and strategies in other countries were based almost exclusively on patient care, in some cases even explicitly excluding research questions. Since 2016, the German Federal Ministry of Education and Research (BMBF) has been supporting the German MII. The objective of the MII is to make data from patient care accessible for medical research, to strengthen data sharing across locations and to establish the necessary infrastructures for this purpose [85], [86]. In a nationwide project framework designed to last a decade (2016–2025), the first phase will focus on university hospital in-patient medicine: Almost all German university medicine locations participate in this initiative and are setting up Data Integration Centers (DICs), which take over the tasks of data extraction from the primary systems, annotation, processing and data provision. In four consortia they are working on various scientific focal issues and use cases [87], [88], [89], [90], but are also jointly working on a nationwide cascaded infrastructure, which includes a standardized patient informed consent and terms of use, as well as a central process for data application in federated data management [85].
The impulse on standardization is based on the fact that a common core data set is to be defined and set up for all university medicine locations. This will be successively harmonized on a module basis and made available at all participating locations [91]. HL7 FHIR has been designated by the MII’s National Steering Committee as the standard to be used for implementing the MII core data set. ART-DECOR is to be used as the tool for dataset modeling and Simplifier/Forge as the tool for creating and publishing FHIR profiles.
Laboratory data are an important basic module of the core data set. The National Steering Committee has stipulated the use of LOINC and UCUM for this purpose. The laboratory analytes – which previously were indeed digital and structured, but coded for the specific location and laboratory – could now be standardized and consolidated across locations. This should make laboratory data available for later research analyses, along with analysis names with unique LOINC ID, examination date, measurement value of the examination with standardized unit, scale type and information on the reference range and interpretation of findings.
This standardization should not be carried out post hoc in the data integration centers’ research database, but preferably in the primary systems. The lab data appear to be particularly suitable for use as an starting point to the harmonization of data sets for data integration, because (a) they are collected on a broad basis from virtually every hospitalized patient, (b) are present in already structured form and (c) are essentially obtained by means of apparatus, that is, the introduction of nomenclatures in the data collection does not interfere with the working and documentation processes by the medical staff [91].
However, the university hospital laboratories in particular have a very large repertoire of analyses performed (up to 12,000 different analytes), of which a large number are only rarely requested and performed and in addition frequently require complicated mapping to LOINC codes. Mapping the entire repertoire to LOINC would therefore require laboratories to expend a lot of effort, which would be countered by limited usefulness: Vreeman et al. were able to show, as early as 2007 in the US – based on an analysis of the laboratory outcome determinations at five different laboratories – that only 2% of the total examination types in a laboratory’s repertoire accounted for 80% of the tests carried out and 20% of the examination types already accounted for 99% of the determinations [92]. This suggests that the entry obstacles to LOINC mapping be lowered in the laboratories and that matters should start with a subset where the scope of analytes to be mapped is limited, but the coverage already exceeds 80%.
This is the principle followed by the MII in Germany: It was decided to start the mapping at all university hospitals with a “LOINC TOP 300” subset, which should already exceed beyond the 80% proportion in German university hospitals. The determination of this subset, which was to comprise 300 types of determination, was carried out by a preliminary analysis at five locations; this identified and compared their most frequent determinations together with their respective proportions in the total number of analyses. In case of variances in the scale type and units or in the system being examined, all relevant codes in the subset were included, so that, per determination, a total of somewhat more than 300 LOINC codes are contained in the TOP 300 subset. Details on the quantity analysis and the definition of the subset will be published soon; data sharing and initial cross-location analysis by means of these data sets (laboratory data module and other basic modules of the core data set, such as diagnoses, procedures, medications) of all university hospitals are scheduled for 2021.
It was possible to expand this standardization initiative beyond the MII at an early stage: In addition to the standardization organizations (HL7 Germany, IHE Germany), actors from patient care (professional associations, specialist societies, the federal institute DIMDI, the KBV) were won over to cooperation, which on its side is currently defining the contents of the state electronic patient records on a statutory basis. By means of cooperation among the responsible institutions in patient care and medical research, there is as great an opportunity to make significant progress in the standardization of laboratory findings data and in the use of LOINC in Germany as there has been in the past almost 20 years.
Perspective for medical research and biobanking
The use of uniform identifiers for laboratory determinations on the basis of internationally agreed standards is of considerable importance for medical research:
Laboratory data is highly relevant for a variety of research questions.
Especially for larger, cross-location research projects and multi-center studies, it is necessary to consolidate the laboratory diagnostic data from different data sources. For this merging a uniform identification of mergeable analytes is necessary.
In the context of research, cross-border use and international evaluation of data is even more relevant than in patient care. Therefore, an internationally uniform identifier is required for laboratory data.
These fundamental concerns of medical research are experiencing a new dynamic through two developments: big data and FAIR explanation follows in detail in the next section.
New high-throughput mass data-processing techniques have created big data technologies capable of linking and analyzing large and heterogeneous amounts of data, partly in real-time applications, and thus providing a new basis for artificial intelligence (AI) applications. Big data technologies that make the data processable will therefore increase the need to combine data from different data sources. However, one of the insights gained from the use of first industrial products (for example, IBM Watson) is the fact that big data technology and AI by no means make the standardization of data – and indeed both their semantic content and technical interfaces – completely superfluous. Also, the problem of the quality of unstructured data for AI has generally been underestimated [93]. Structuring, quality improvement and standardization of clinical data, especially including valuable laboratory diagnostic data, are therefore a prerequisite for the success of AI and big data technologies – the demand will increase.
In addition, in research worldwide and across domains, the idea of enabling more reuse of research data is also gaining ground – on the one hand to achieve better data quality and traceability (through professionalized data management and data stewardship), and on the other hand to use research resources more sparingly. The focus here is on addressing a reuse of data stocks that is “machine actionable”, especially in view of increasing data volumes (big data). The international GO-FAIR initiative has developed the so-called FAIR criteria, which every scientist should take into account for the research data stocks that he has set up and uses. Specifically, data sets should be F: findable, A: accessible, I: interoperable and R: reusable [94], [95]. Within a few years, the FAIR criteria have become a quasi-self-commitment in science. It is obvious that more reuse of research data increases the requirement for more standardization and that interoperability is already entrenched as an important prerequisite in the criteria. With regard to medical laboratory diagnostic data, this too will contribute significantly to the need for standardization – which ultimately always has to be implemented at the data source, i.e. in the laboratory. Obviously, it is not enough to have uniform identifiers for laboratory test and their units (by means of LOINC and UCUM): Non-quantitative results must also be expressed in a standardized way (by means of SNOMED CT). Furthermore, provenance metadata and information for assessing data quality must be recorded and carried along, for example, information on the context in which data is obtained, quality assurance steps carried out or data transformations that occur. It may also be necessary to code diagnostic processes at below the METHOD level in LOINC to ensure comparability and reusability. Overall, even for a long-established and largely unified field such as laboratory medical diagnostics, this is not a trivial undertaking, but rather a complex information model and data communication process, for which it is still unclear who bears the costs for the quality-related data collection and management on a sustainable basis. In Germany, not only the MII that is working on these issues of the future, but also the National Research Data Infrastructure (NFDI), a large-scale infrastructure funding measure of the Federal Government and the German Federal States that will be launched in 2020 and will cover all research areas.
Finally, there is another specific challenge for German health research: the German health care system is traditionally fragmented into different sectors, within which actors for the service providers and funding carriers are responsible according to the principle of self-management, while the federal and state health ministries are only indirectly responsible on many issues [96], [97]. The data sets of the German health care system follow this structure and – unlike in other European countries with strong central structures (such as the UK) or with several central patient registers (such as the Scandinavian countries) – are distributed sectorally and federally and are highly fragmented [98], [99], [100]. Thus, the data stocks in the out-patient and in-patient sectors, in rehabilitation facilities or in federal offices are by no means linked or comparable. Necessary information from these data sets, which are particularly relevant for health service research [98], [101], must be painstakingly brought together and consolidated. In addition to the legal obstacles of data protection and the technical obstacles of record linkage, the lack of data standardization also emerges as a problem. Standardization (e.g. with LOINC for the lab data) in only one sector or partial data set has only a limited effect; therefore, nationally coordinated measures are needed to achieve the objective of increasing the use of existing data in the German health care system for medical research.
Laboratory data standardization is of potential interest also for biobanks. However, the subject of LOINC in biobanking has not yet been elucidated in the literature. Biobanks, the “biological back end of data-driven medicine” [102], are essentially research infrastructures that store samples and make them available for research inquiries that are linked to clinical data to a varying extent. Quality assurance and standardization are therefore necessary not only in the area of pre-analytics, analytics and storage [103], but also for data processing. IT interoperability is at the same time required for various aspects of biobanking: sample-related data, clinical annotation data, control for sample processing and storage, handling of informed consent and feedback processes [104].
Thus far, European efforts have focused in particular on standardizing the questions of: (a) how a biobank describes itself and its sample collections, in order to make them findable, for example in national or international registers, and (b) how the individual sample collections can be characterized in such a way as to enable the identification of samples for subsequent use for specific inquiries. The first aspect has been successfully resolved internationally by the European data set coordinated by the BBMRI, Minimum Information About BIobank Data Sharing (MIABIS). A second version of MIABIS has recently been published, which makes it possible to describe biobanks, sample collections and individual studies in a biobank in a modular way [102], [105]. The second aspect is more complex. The task here is to define a standardized, searchable data set from the phenotypic data assigned to each sample, which contains the relevant criteria for searching for a sample. In particular diagnoses in ICD-coded form or, in tissue samples, information from the pathology are applied here. Laboratory data, on the other hand, are usually not used, even if these in some cases are pathognomonic and in addition represent frequent inclusion and exclusion criteria for clinical trials. As far as a biobank is not located close to the health care provider (health care-integrated biobanking) and is directly connected to a laboratory information system (LIS), these data are often not available at all.
In principle, laboratory data would be relevant as part of the annotation of samples with phenotypic clinical data. Since the biobanks act both as receivers and as senders in distributed systems and networks, LOINC standardization would be valuable in several scenarios (see Figure 5): 1. A biobank receives samples and data from multiple sources. Standardization is warranted for internal consolidation – in addition to ICD-encoded diagnoses, LOINC-encoded laboratory results from the clinical data on the patient or sample donor could contribute to characterizing the sample. This information can then be used in search queries for specific samples. 2. The biobank in turn provides samples and clinical data to scientists. If it has received and retained the clinical data, including the characteristic laboratory data, it can also provide it. Since receivers are likely to have problems with proprietary identifiers in automated further processing, the provision of LOINC standardization would be valuable. 3. The biobank itself acts as an analytics provider and sends newly acquired analysis data (with or without a sample) from the sample to scientists who will reuse it. Here also, the receiver benefits from standardization in further processing; here, the analysis facilities of the biobank or its “Biomaterial Information and Management System” (BIMS) would have to label the data with LOINC codes itself and maintain this coding and then provide it as well. 4. A further case for application concerns health care-integrated biobanking: in clinics, many samples are accumulated in the course of the treatment process as left-over samples, and, beyond dedicated studies, it is necessary to identify which sample is of interest to the biobank according to appropriate criteria. This must be done using information obtained as early as possible during the patient’s stay. In addition to the admission diagnosis, here usually early laboratory data from the panel diagnostics are particularly appropriate parameters. Since such a selection must be made algorithmically due to the high sample volume, the criteria must be available in structured and standardized form – the latter in particular if selection is to be made according to uniform criteria at different locations and from different laboratories. Here, too, standardization by means of LOINC offers a suitable basis.
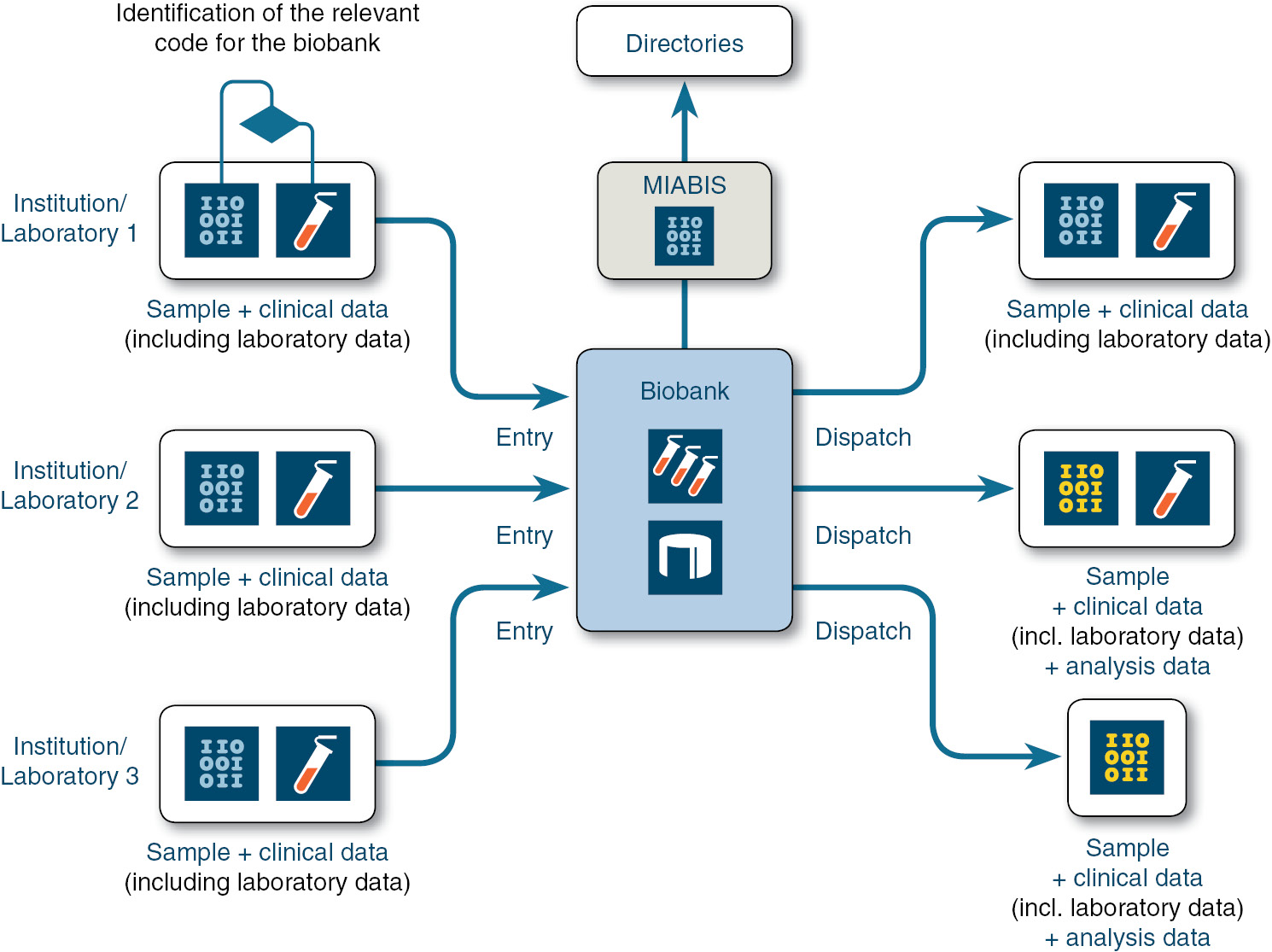
Biobanks as recipients and senders of information that also includes laboratory data.
The use of LOINC to identify analyses can contribute to interoperability also in these scenarios.
All the benefits of standardizing information in the clinical data available to biobanks naturally apply equally to other data types and standards. Of course, laboratory data are particularly relevant (also for one’s own analysis), are already structured and are available on a large scale for every treatment case, and standardization with LOINC is comparatively simple, so this may be an appropriate first step for making clinical data accessible to biobanks. More complex standardizations, such as for OMICS data, will also be necessary. Overall, biobanks should therefore be seen and involved as actors in digitization, and the BIMS should be part of a higher-level IT architecture, since links to other data bases will be required in both directions: On the one hand, the access to linked data sets, for example in an electronic patient record on the donor of a sample will also be relevant for the biobank; on the other hand, the analysis data generated from a biosample will in turn also have to be available for the IT systems for patient care, medical research and, last but not least, for the patient. Of course, there are many legal and technical challenges ahead for such system and data integration [106].
A certain amount of standardization has already been done for sample-related data, namely by SPREC, which describes the pre-analytical situation of a sample and has been internationally reconciled by the International Society for Biological and Environmental Repositories (ISBER). First issued in 2010 and updated in a first revision in 2012, the third version of SPREC was published in 2018 [107], [108], [109].
SPREC expresses the essential critical pre-analytical variables for fluid and tissue samples in a relatively simple composed code. Table 3 and Figure 6 show the seven elements of the code system, which are expressed in one code for a sample, using different numbers of digits per code element, and thus identify the pre-analytical characteristics of the sample in a standardized and automated machine-readable manner. A relatively manageable pre-coordinated value set is defined for each of the seven elements; this contains the approved values and code digits and is oriented to the relevant laboratory medical and clinical pathological conditions. The number of possible values as well as the definition of the values in detail has changed and has been further developed over the previous versions of SPREC. Table 3 shows the respective number of allowed values per element, based on SPREC Version 3.0 of 2018 [109].

Example of a SPREC code and its composition.
SPREC code system, for fluid and tissue samples.
| Fluid biospecimen | Solid biospecimen | ||||
|---|---|---|---|---|---|
| 11 digits | 13 digits | ||||
| 7 elements | 7 elements | ||||
| Element | No. of values | Code digits | Element | No. of values | Code digits |
| Type of sample | 36 | 3 digits | Type of sample | 13 | 3 digits |
| Type of primary container | 35 | 1 digit | Type of collection | 18 | 3 digits |
| Precentrifugation (delay between collection and processing) | 19 | 1 digit | Warm ischemia time | 9 | 1 digit |
| Centrifugation | 14 | 1 digit | Cold ischemia time | 17 | 1 digit |
| Second centrifugation | 13 | 1 digit | Fixation/stabilization type | 15 | 1 digit |
| Postcentrifugation delay | 14 | 1 digit | Fixation time | 11 | 1 digit |
| Long term storage conditions | 26 | 1 digit | Long term storage conditions | 26 | 1 digit |
| SPREC code syntax: Elements 1–7, hyphen-separated. Standardized vocabulary (value set) | |||||
The additional columns show the number of digits in the code as well as the number of values allowed per element in the respective value set (SPREC Version 3.0, 2018).
These SPREC codes can be generated automatically, at least in part, from procedural protocols and documentation in the laboratory or pathology.
If a biobank, a clinical laboratory or a pathology institute wants to use LOINC and SPREC simultaneously, note should be taken of a semantic overlap between both nomenclatures: the TYPE OF SAMPLE element in SPREC, just like the SYSTEM axis in LOINC, represents information on the type of sample being examined. Unfortunately, there is no easy way of simple assignment here, since one or the other nomenclature describes the sample type in a more detailed and granular way (n:m relationship). Some examples:
“Amniotic fluid [AMN]” (SPREC TYPE OF SAMPLE) clearly corresponds to “Amnio fld” (LOINC SYSTEM). The same applies to “Tears [TER]” (SPREC) vs. “Tear” (LOINC).
“Ascites fluid [ASC]” (SPREC TYPE OF SAMPLE) can at least likewise be reciprocally and unambiguously assigned to “Periton fld” (LOINC SYSTEM) – even though here SPREC addresses only one sub-entity, the pathological accumulation of peritoneal fluid.
For “Stool [STL]” (SPREC), however, LOINC differentiates between “Stool”, “Stool.wet” and “Stool.dried”. The same applies for “Serum [SER]”, which SPREC only recognizes in a general form, whereas LOINC recognizes at least 12 different forms, including residual classes to identify unknown differentiation.
For urine samples, the situation is the other way around: LOINC SYSTEM only recognizes “Urine” (except for the additional identification of fetal urine, “Urine^fetus”), while SPREC here already distinguishes among “Urine, random (“spot”) [URN]”, “Urine, first morning [URM]” and “Urine, timed [URT]”.
Considered in detail, here SPREC deviates in its differentiation from a pure description of the starting material and already includes processing steps or the intentions of the examination. This is also illustrated by the example of plasma: LOINC System recognizes “Plas” and – if serum/plasma cannot or does not need to be differentiated any more clearly – “Ser/Plas”. SPREC, on the other hand, makes an obligatory differentiation according to the centrifugation state of the plasma, between “Plasma, single spun [PL1]” or “Plasma, double spun [PL2]”.
Assignment becomes more complex because LOINC regards these elements (not always completely consistently, but in principle logically) as components of the examination, not of the examined sample material, and thus expresses in its COMPONENT axis.
A professional, quality-assured mapping would be desirable, ideally centrally maintained in a coordinated manner (or taken into account in the future development of SPREC or LOINC), in order to be able to reciprocally support an automatable generation of SPREC and LOINC codes or partial elements of the respective code systems.
In principle, SNOMED CT could be used for the transfer of one into the other, which could also fully map SPREC. However, there is a need for simple code systems in specialist sub-domains, such as biobanking, that can be managed without an IT superstructure.
Conclusion and outlook
Twenty-five years after the first publication of LOINC, the standardization of lab data will first become really urgent. Data integration will not only be the promise, but also the challenge if the coming years. Increasingly, citizens and patients themselves are becoming addressees of such offerings, now that platforms are available for them that not only generate data using smartphone-based sensor technology, but also integrate app-based data and prepare it for the citizen. The new statutory requirements for a comprehensive electronic patient record for citizens in Germany will bring new opportunities for, increased interest in and higher demands on the longitudinal consolidation, processing and presentation of citizens’ health data. Such a longitudinal view will provide opportunities to close gaps between practitioners and to support treatment on the basis of data. The evaluation of longitudinal profiles also offers new possibilities for research, such as case-based reasoning and disease modeling. At the same time, new questions will arise in the course of longitudinal merging of health data in the citizen’s apps or EPR: How comparable are the contents of test results from different laboratories, analytics providers and sensors – which can be consolidated thanks to LOINC- and UCUM-based standardization? Which interpretation aids must be provided to the citizen for considering such data consolidations? For networked and citizen-oriented digital medicine, laboratory data offer great potential, but also challenges to be solved [110].
Nevertheless, the following applies: Without standardization, such a longitudinal consolidation will not succeed. It will become increasingly important that standardization will be considered not only for individual core data sets or partial data stocks, but also from a holistic approach, and matters will focus in particular on those data that are particularly relevant to everyday life for EPRs and citizens’ apps. In addition to diagnoses and laboratory data, this includes, in particular, medication data, the standardized documentation of which is still considerably lacking in Germany, but where international experience with interaction and convergence of various semantic standards is certainly available [111]. SNOMED CT will play an important role here, even though not all problems will be solved by the national introduction of SNOMED CT. Besides this, Germany is not yet a member of SNOMED International and therefore not authorized to implement SNOMED CT. At the end of 2019, the Federal Ministry of Education and Research (BMBF) will initiate access to the use of SNOMED CT in Germany via the MII. Nevertheless, it is a pragmatic approach to use individual domain-specific nomenclatures (such as LOINC for laboratory data), if necessary only for certain defined parts of the documentation, in order to achieve greater acceptance and dissemination. A “one size fits all” approach would likely fail. Rather, it seems important to keep entry barriers low and technical complexity manageable. A start-up done on too large a scale will hinder taking the important small first steps. In this respect, the 1994 statement of the AMIA Board of Directors, at the beginning of the development of LOINC, should be cited once again: “The goal is to have an acceptable code system for each kind of data. It is not necessary (it may not even be desirable) to have all of the codes come from a single master code system, because computers can integrate multiple code systems easily while avoiding collisions among assigned codes by adding a code source designation. Consequently, we can create a suitable first-phase set of codes for the computer-based medical record by borrowing from many different existing code systems” [16]. These guiding principles are also valid and relevant for 2019 – including for Germany and for harmonization in the European Union.
GLOSSARY
- AG
Arbeitsgruppe (working group)
- AI
Artificial Intelligence (in German: Künstliche Intelligenz = KI)
- AIM
Advanced Informatics in Medicine, historical EU funding programme 1988–1994 within the EU Third Framework Programme
- AMA
American Medical Association (www.ama-assn.org) AMIAAmerican Medical Informatics Association (www.amia.org)
- AMTS
Arzneimitteltherapiesicherheit [Drug therapy safety]
- ANSI
American National Standards Institute (www.ansi.org)
- ART-DECOR
Advanced Requirement Tooling – Data Elements, Codes, OIDs and Rules: open-source tool suite that supports the creation and maintenance of HL7 templates, value sets, scenarios and data sets O (https://art-decor.org)
- ASTM
American Society for Testing and Materials (www.astm.org)
- ATC
Anatomical Therapeutic Chemical (ATC) Classification System, provided since 1976 by the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC)
- BBMRI
[European] Biobanking and Biomolecular Resources Research Infrastructure, a ESFRI unit within the EU (www.biobanks.eu)
- BIMS
Biomaterial Information and Management System (Software)
- BITKOM
Bundesverband Informationswirtschaft, Telekommunikation und neue Medien e.V. [Federal Association for Information Technology, Telecommunications and New Media in Germany] (www.bitkom.org)
- BMBF
Bundesministerium für Bildung und Forschung [Federal Ministry of Education and Research] (www.bmbf.de)
- BMG
Bundesministerium für Gesundheit [Federal Ministry of Health] (www.bmg.bund.de)
- bvitg
Federal industrial association of Health IT in Germany (www.bvitg.de)
- CCR
Continuity of Care Record, a core data set & patient summary standard, provided by the American Society for Testing and Materials (ASTM) (www.astm.org/Standards/E2369.htm)
- CDA
Clinical Document Architecture, HL7 standard for the exchange of clinical documents
- CDI
Clinical data interchange
- CDISC
Clinical Data Interchange Standards Consortium, a standards developing organization (SDO) and name of a family of data standards for clinical research (www.cdisc.org)
- CEN
Comité Européen de Normalisation [European Committee for Standardisation] (www.cenorm.be)
- C-NPU
Committee on Nomenclature, Properties and Units (www.ifcc.org/divisions/sd/c-npu.asp)
- conhIT
Connecting Healthcare IT; industrial fair for information technology in the healthcare sector in Germany; 2018 rebranded to “DMEA”
- CPT
Current Procedural Terminology, a medical code set in the USA, since 1966 developed and provided by the American Medical Association (AMA)
- CSV
Comma Separated Values
- CT
Controlled Terminology
- CUMUL
name [no acronym] of a historical medical informatics project in Switzerland 1997–2001 by the Centre Suisse de Contrôle de Qualité (CSCQ) [Quality Control Center Switzerland] at Geneva (formerly available at www.cumul.ch)
- DFG
Deutsche Forschungsgemeinschaft [German Research Foundation], the central, independent and self-governing research funding organisation in Germany (www.dfg.de)
- DIC
Data Integration Center
- DICOM
Digital Imaging and Communications in Medicine (http://medical.nema.org)
- DIMDI
Deutsches Institut für Medizinische Dokumentation und Information (German Institute of Medical Documentation and Information [DIMDI]), a federal agency and authority within the German Federal Ministry of Health (www.dimdi.de)
- DIN
Deutsches Institut für Normung e.V. [German Institute for Standardisation] in Germany (www.din.de)
- DMEA
DMEA – Connecting Digital Health: industrial IT fair in Germany (www.dmea.de)
- DVG
Gesetz für eine bessere Versorgung durch Digitalisierung und Innovation (Digitale-Versorgung-Gesetz – DVG) [Act to Improve Healthcare Provision through Digitalisation and Innovation – Digital Health Care Act] in Germany (2019)
- EDIFACT
Electronic Data Interchange for Administration, Commerce and Transport; a global, cross-sector message standard for electronic data exchange since 1886, provided by the United Nations Centre for Trade Facilitation and Electronic Business (CEFACT)
- EHG
E-Health Act in Germany (2015)
- EHR
Electronic Health Record
- ELGA
ELektronische Gesundheitsakte [Electronic Health Record] in Austria (www.elga.gv.at)
- ENV
draft European standard of CEN
- EPR
Electronic Patient Record
- epSOS
European Patients Smart Open Services; EU project to promote interoperability of eHealth systems for emergency data and electronic prescriptions (www.epsos.eu)
- ESFRI
European Strategy Forum on Research Infrastructures (http://cordis.europa.eu/esfri)
- EUCLIDES
European Clinical Laboratory Data Exchange Standard, historical EU project and IT standard for lab data exchange (1988 – ca. 1995)
- FAIR
Findable, Accessible, Interoperable, Reusable: principles of the international GO FAIR initiative for better research data (www.go-fair.org)
- FAQ
Frequently asked question(s)
- FB
Fachbereich [Department]
- FDA
US Food and Drug Administration (www.fda.gov)
- FHIR
Fast Healthcare Interoperability Resources; a HL7 standard (http://hl7.org/fhir)
- Gematik
Gesellschaft für Telematikanwendungen der Gesundheitskarte mbH [company for telematic applications of the health card Ltd.], agency for health telematics in Germany (www.gematik.de)
- GFISCO
GO FAIR International Support and Coordination Office (www.go-fair.org/go-fair-initiative/go-fair-offices/)
- GMDS
Deutsche Gesellschaft für Medizinische Informatik, Biometrie und Epidemiologie e.V. [German Society for Medical Informatics, Biometry and Epidemiology] (www.gmds.de)
- GMS
German Medical Science web portal (www.egms.de)
- GO FAIR
international initiative for better research data (www.go-fair.org)
- HL7
Health Level Seven; international Standards Development Organisation in the area of interoperability of healthcare IT systems (www.hl7.org)
- ICD
International Statistical Classification of Diseases and Related Health Problems
- ICH
International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (www.ich.org)
- ID
Identification number
- IEEE
International non-profit organization and professional association for the advancement of technology, originally: Institute of Electrical and Electronics Engineers (www.ieee.org)
- IFCC
International Federation of Clinical Chemistry and Laboratory Medicine (www.ifcc.org)
- IHE
Integrating the Healthcare Enterprise (www.ihe.net)
- IHTSDO
International Health Terminology Standards Development Organisation (www.ihtsdo.org); international non-profit organisation, trading as SNOMED International (www.snomed.org)
- ISBER
International Society for Biological and Environmental Repositories (www.isber.org)
- ISO
International Organization for Standardization (www.iso.org)
- ITeG
IT-Messe im Gesundheitswesen [IT industrial fair in the healthcare sector in Germany]; 2008 rebranded to “conhIT” and 2018 rebranded to “DMEA”
- IUPAC
International Union of Pure and Applied Chemistry (www.iupac.de)
- IUSM
Indiana University School of Medicine, Indianapolis, United States (www.medicine.iu.edu)
- KBV
Kassenärztliche Bundesvereinigung [German Federal Association of Statutory Health Insurance Physicians] (www.kbv.de)
- LAB
Laboratory Data Model (CDISC-Standard)
- LB
Laboratory Tests; Finding Domain in CDISC-SDTM
- LDT
Labordatenträger [Lab Data Media], a standard for data exchange between laboratory and practice software systems for laboratory data in Germany, provided by the German Federal Association of Statutory Health Insurance Physicians [Kassenärztliche Bundesvereinigung, KBV]
- LIS
Laboratory Information System (Software)
- LOINC
Logical Observation Identifiers Names and Codes (www.loinc.org)
- MDR
Metadata Repository
- MedDRA
Medical Dictionary for Regulatory Activities (www.meddra.org)
- MIABIS
Minimum Information About BIobank data Sharing (Standard of BBMRI)
- MII
Medizininformatik-Initiative [Medical Informatics Initiative Germany, funded by the Federal Ministry of Education and Research, BMBF] (www.medizininformatik-initiative.de)
- MWV
Medizinisch Wissenschaftliche Verlagsgesellschaft OHG, Berlin [Medical Scientific Publishing Company] (www.mwv-berlin.de)
- NA
Normenausschuss des DIN [Standards Committee of the German Institute for Standardization, DIN]
- NAMed
NA 063 Normenausschuss Medizin des DIN [Standards Committee for Medicine of the German Institute for Standardization, DIN] (www.named.din.de)
- NFDI
Nationale Forschungsdateninfrastruktur [National Research Data Infrastructure, in Germany, starting in 2020]
- NHS
UK National Health Service (www.nhs.uk)
- NIH
US National Institutes of Health (www.nih.gov)
- NPU
Nomenclature for Properties and Units, a clinical laboratory terminology for use in the clinical laboratory science (www.npu-terminology.org)
- OID
Object Identifier
- openEHR
open standard specification for health data in electronic health records (EHRs), maintained by the openEHR Foundation (www.openehr.org) and built upon the basis work of the European AIM project Good European Health Record (GEHR) (1989-1994)
Portable Document Format [developed by Adobe Inc.] (www.adobe.com)
- PDMS
Patient data management system in Anesthesiology and Intensive Care Medicine
- RELMA
Regenstrief LOINC Mapping Assistant
- SDO
Standards Development Organisation
- SDTM
Study Data Tabulation Model (CDISC-Standard)
- SHI
Statutory Health Insurance (in Germany)
- Simplifier
SIMPLIFIER.NET: FHIR Collaboration Platform (https://simplifier.net)
- SNOMED CT
Systematized Nomenclature of Medicine – Clinical Terms (www.snomed.org)
- SNOMED
Systematized Nomenclature of Medicine (www.snomed.org)
- SPREC
Standard PREanalytical Code [of the Biospecimen Science Working Group of ISBER]
- STM
Standardisierte Terminologien in der Medizin; Projektgruppe der GMDS [Standardized Terminologies in Medicine; project group oft he GMDS] (www.imi.uni-luebeck.de/gmds-ag-stm)
- TC
Technical Committee
- TMF
TMF – Technologie- und Methodenplattform für die vernetzte medizinische Forschung e.V. [TMF – Technology, Methods, and Infrastructure for Networked Medical Research], NPO for digitalisation in medical research in Germany (www.tmf-ev.de)
- TSVG
Gesetz für schnellere Termine und bessere Versorgung – Terminservice- und Versorgungsgesetz [Appointment Service and Health Care Act] (2019)
- UCUM
Unified Code for Units of Measure (http://unitsofmeasure.org)
- VDAP
Verband deutscher Arztpraxis-Softwarehersteller e.V. [Association of German medical practice software manufacturers] (www.vdap.de)
- VHitG
obsolete: Verband der Hersteller von IT-Lösungen für das Gesundheitswesen e.V. [Association of manufacturers of IT solutions for the healthcare industry]; 2011 renamed to bvitg e.V. (www.bvitg.de)
- VITAL
Vital Signs Information Representation, a historical CEN standard
Author contributions: The author has accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding: None declared.
Employment or leadership: None declared.
Honorarium: None declared.
Competing interests: The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
References
1. LOINC Committee, Regenstrief Institute. http://www.loinc.org. Accessed: 6 Oct 2019.Search in Google Scholar
2. Forrey AW, McDonald CJ, DeMoor G, Huff SM, Leavelle D, Leland D, et al. Logical observation identifier names and codes (LOINC) database: a public use set of codes and names for electronic reporting of clinical laboratory test results. Clin Chem 1996;42:81–90.10.1093/clinchem/42.1.81Search in Google Scholar
3. McDonald CJ, Park BH, Blevins L. Grocers physicians and electronic data processing. AMA Cont Med Ed Newsletter 1983;12:5–8.Search in Google Scholar
4. McDonald CJ. Standards for the electronic transfer of clinical data: progress, promises, and the conductor’s wand. Proc Annu Symp Comput Appl Med Care 1990:9–14.Search in Google Scholar
5. McDonald CJ. Standards for the transmission of diagnostic results from laboratory computers to office practice computers – an initiative. Proc Annu Symp Comput Appl Med Care 1983:123–4.10.1109/SCAMC.1983.764563Search in Google Scholar
6. ASTM E1238-88 Standard Specification for Transferring Clinical Laboratory Data Messages Between Independent Computer Systems, 1988.Search in Google Scholar
7. Huff SM. Clinical data exchange standards and vocabularies for messages. Proc AMIA Symp 1998:62–67.Search in Google Scholar
8. Spronk R. The early history of health Level 7. Ringholm Whitepaper 2014. http://www.ringholm.com/docs/the_early_history_of_health_level_7_HL7.htm. Accessed: 6 Oct 2019.Search in Google Scholar
9. McDonald CJ, Martin DK, Overhage JM. Standards for the electronic transfer of clinical data: progress and promises. Top Health Rec Manage 1991;11:1–16.Search in Google Scholar
10. Rossing N. AIM: advanced informatics in medicine the AIM exploratory action 1988–1990. In: van Goor JN, Christensen JP, editors. Advances in medical informatics – results of the AIM exploratory action. Amsterdam: IOS, 1992:1–4.Search in Google Scholar
11. Schug SH. European and international perspectives on telematics in healthcare. Köln: GVG/Aka, 2001.Search in Google Scholar
12. France FR. About the beginnings of medical informatics in Europe. Acta Inform Med 2014;22:11–15.10.5455/aim.2014.22.11-15Search in Google Scholar PubMed PubMed Central
13. De Moor GJ. EUCLIDES: A European clinical laboratory data exchange standard (AIM-project: A 1033). In: Adlassnig KP, Grabner G, Bengtsson S, Hansen R, editors. Medical Informatics Europe 1991. Lecture Notes in Medical Informatics. Berlin, Heidelberg: Springer, 1991:876–7.10.1007/978-3-642-93503-9_156Search in Google Scholar
14. De Moor GJ. EUCLIDES: a European clinical laboratory data exchange standard (AIM-Project: A 1033). In: van Goor JN, Christensen JP, editors. Advances in Medical Informatics – Results of the AIM Exploratory Action. Amsterdam: IOS, 1992:108–16.Search in Google Scholar
15. De Moor GJ. Towards a standard for electronic data interchange in laboratory medicine. Ghent: University of Ghent, 1994.Search in Google Scholar
16. Board of Directors of the American Medical Informatics Association (AMIA). Standards for medical identifiers, codes, and messages needed to create an efficient computer-stored medical record. American Medical Informatics Association. J Am Med Inform Assoc 1994;1:1–7.10.1136/jamia.1994.95236133Search in Google Scholar PubMed PubMed Central
17. Huff SM, Rocha RA, McDonald CJ, De Moor GJ, Fiers T, Bidgood WD, et al. Development of the logical observation identifier names and codes (LOINC) vocabulary. J Am Med Inform Assoc 1998;5:276–92.10.1136/jamia.1998.0050276Search in Google Scholar PubMed PubMed Central
18. McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 2003;49:624–33.10.1373/49.4.624Search in Google Scholar PubMed
19. Semler SC, Röhrig R. LOINC – Internationale Nomenklatur zur Kodierung von medizinischen Untersuchungen und Befunden. In: Rienhoff O, Semler SC, editors. Terminologien und Ordnungssysteme in der Medizin. Berlin: MWV, 2015:97−134.10.32745/9783954665174-2.7Search in Google Scholar
20. Cooperation Agreement, dated July 2013, between The International Health Terminology Standards Development Organisation (IHTSDO) and The Regenstrief Institute, Incorporated (RII). https://loinc.org/collaboration/ihtsdo/agreement.pdf. Accessed: 6 Oct 2019.Search in Google Scholar
21. McDonald CJ, Huff S, Deckard J, Holck K, Abhyankar S, Vreeman DJ. Logical Observation Identifiers Names and Codes (LOINC®) Users’ Guide – June 2015, Regenstrief Institute, Inc. and the Logical Observation Identifiers Names and Codes (LOINC) Committee. https://loinc.org/downloads/files/LOINCManual.pdf. Accessed: 6 Oct 2019.Search in Google Scholar
22. Vreeman DJ. LOINC Essentials – a step by step guide to getting your local codes mapped to LOINC. Blue Sky Premise, 2016. https://danielvreeman.com/loinc-essentials/.Search in Google Scholar
23. Heubeck C, Fiebrig I, Criegee-Rieck M. LOINC Benutzerhandbuch. (Dt. Übersetzung von: McDonald C, Huff S, Vreeman DJ, Mercer K. LOINC User Guide.) DIMDI, 2005.Search in Google Scholar
24. Semler SC. Standardisierte Labor- und Vitaldatenkommunikation: LOINC – die unbekannte Norm in Deutschland. In: Wehrs H, editor. Der Computer-Führer für Ärzte – Ausgabe 2003. Dietzenbach: Antares, 2002:78–85.Search in Google Scholar
25. Schadow G, McDonald CJ, Suico JG, Fohring U, Tolxdorff T. Units of measure in clinical information systems. J Am Med Inform Assoc 1999;6:151–62.10.1136/jamia.1999.0060151Search in Google Scholar PubMed PubMed Central
26. Schadow G, McDonald CJ. UCUM Webseite. http://www.unitsofmeasure.org/. Accessed: 6 Oct 2019.Search in Google Scholar
27. Semler SC. Labor- und Vitaldatenkommunikation mittels LOINC – erste Erfahrungswerte aus Implementationen in Deutschland. In: Steyer G, et al., editors. TELEMED 2002: Tagungsband zur 7. Fortbildungsveranstaltung und Arbeitstagung. Berlin, 2002:213–23.Search in Google Scholar
28. Semler SC. Labor- und Vitaldatenkommunikation – Standardisierung durch LOINC. Dt. Ärzteblatt 2003;100(Suppl):10–13.Search in Google Scholar
29. Weigand C. VITAL: use and implementation of a medical communication standard in practice. Comput Cardiol 2005;32:319–22.10.1109/CIC.2005.1588101Search in Google Scholar
30. Benson T. The history of the Read Codes: the inaugural James Read Memorial Lecture 2011. Inform Prim Care 2011;19:173–82.10.14236/jhi.v19i3.811Search in Google Scholar PubMed
31. Cormont S, Erman A, Burckel Y, Carayon A. Names-Lab®: un modèle de normalisation des messages de biologie. Ann Biol Clin (Paris) 2002;60:173–81.Search in Google Scholar
32. Gonzalez Gacio G, Cilla Eguiluz CC, Lazaro Naranjo MA, Berez Baldrich MV, Rodriguez Naque M, Mota AM, et al. Adoption of clinical vocabularies LOINC® and SNOMED CT® in the microbiology laboratory and their integration in the information systems and electronic medical records. Clin Chima Acta 2019;493(Suppl 1):S110–1.10.1016/j.cca.2019.03.234Search in Google Scholar
33. Dybkaer R, Jørgensen K. Quantities and units in clinical chemistry including Recommendation 1966 of the commission on clinical chemistry of the IUPAC and of the IFCC. Copenhagen: Munksgaard; 1967:x–102.Search in Google Scholar
34. Dybkaer R. Quantities and units in clinical chemistry. Clin Chem 1968;14:989–92.10.1093/clinchem/14.10.989Search in Google Scholar
35. Olesen H. Properties and units in the clinical laboratory sciences I. Syntax and semantic rules. Pure Appl Chem 1995;67:1563–74.10.1351/pac199567081563Search in Google Scholar
36. Kenny D, Olesen H. Properties and units in the clinical laboratory sciences II. Kinds-of-property. Pure Appl Chem 1997;69:1015–42.10.1351/pac199769051015Search in Google Scholar
37. Bruunshuus I, Frederiksen W, Olesen H, Ibsen I. Properties and units in the clinical laboratory sciences: part III Elements (of properties) and their code values. Pure Appl Chem 1997;69:2577–82.10.1351/pac199769122577Search in Google Scholar
38. Olesen H, Kenny D, Bruunshuus I, Ibsen I, Jørgensen K, Dybkaer R, et al. Properties and units in the clinical laboratory sciences: part IV. Properties and their code values. Pure Appl Chem 1997;69:2583–91.10.1351/pac199769122583Search in Google Scholar
39. Olesen H, Ibsen I, Bruunshuus I, Kenny D, Dybkær R, Fuentes-Arderiu X, et al. Properties and units in the clinical laboratory sciences. X. Properties and units in general clinical chemistry (Technical Report, IFCC-IUPAC 1999). Pure Appl Chem 2000;72:747–972.10.1351/pac200072050747Search in Google Scholar
40. Olesen H, Kenny D, Dybkær R, Ibsen I, Bruunshuus I, Fuentes-Arderiu X, et al. Properties and units in the clinical laboratory sciences: part XI. Coding systems – structure and guidelines (Technical Report, IUPAC-IFCC 1997). Pure Appl Chem 1997;69:2607–20.10.1351/pac199769122607Search in Google Scholar
41. Petersen UM, Dybkær R, Olesen H. Properties and units in the clinical laboratory sciences. Part XXIII. The NPU terminology, principles, and implementation: a user’s guide (IUPAC Technical Report). Pure Appl Chem 2012;84:137–65.10.1351/PAC-REP-11-05-03Search in Google Scholar
42. Petersen UM, Padró-Miquel A, Taylor G, Hertz JM, Ceder R, Fuentes-Arderiu X, et al. Properties and units in the clinical laboratory sciences part XXIV. Properties and units in clinical molecular genetics (IUPAC Technical Report). Pure Appl Chem 2018;90:1199–220.10.1515/pac-2017-1008Search in Google Scholar
43. Férard G, Dybkaer R, Fuentes-Arderiu X. The silver book and the NPU format for clinical laboratory science reports regarding properties, units, and symbols. Chem Int 2017;39:A1–4.10.1515/ci-2017-0217Search in Google Scholar
44. Pontet F, Magdal Petersen U, Fuentes-Arderiu X, Nordin G, Bruunshuus I, Ihalainen J, et al. Clinical laboratory sciences data transmission: the NPU coding system. Stud Health Technol Inform 2009;150:265–9.Search in Google Scholar
45. Férard G, Dybkaer R. Recommendations for clinical laboratory science reports regarding properties, units, and symbols: the NPU format. Clin Chem Lab Med 2013;51:959–66.10.1515/cclm-2013-5000Search in Google Scholar
46. Külpmann WR. Das NPU-System (IFCC, IUPAC) der Messgrößen und Einheiten in der Laboratoriumsmedizin. J Lab Med 2005;29:2–5.Search in Google Scholar
47. NPU Terminology – Danish Release Center. The Danish Health Data Authority. http://www.npu-terminology.org. Accessed: 6 Oct 2019.Search in Google Scholar
48. IHTSDO, Regenstrief Institute, IFCC, IUPAC. Owners of LOINC, NPU, and SNOMED CT Begin Trial of Cooperative Terminology Development – Joint Press Release. 2009. http://www.nlm.nih.gov/research/umls/Snomed/press_release.html. Accessed: 6 Oct 2019.Search in Google Scholar
49. IFCC, IUPAC, IHTSDO, Regenstrief Institute. Memorandum of Understanding among International Federation of Clinical Chemistry and Laboratory Medicine [IFCC] and the International Union of Pure and Applied Chemistry [IUPAC], International Health Terminology Standards Development Organisation FMBA [IHTSDO] established under the laws of Denmark (CVR 30363434), The Regenstrief Institute, Inc. [RI] Regarding an operational trial of a division of labor in laboratory test terminology development involving LOINC, NPU & SNOMED CT. 2009. http://www.nlm.nih.gov/research/umls/Snomed/loinc_npu_mou.html. Accessed: 6 Oct 2019.Search in Google Scholar
50. de Haan JB, Deom A. Pour un répertoire universel des analyses médicales: LOINC™, version française. Ann Biol Clin 1999;57:730–6.Search in Google Scholar
51. de Haan JB. ELM Cumul Project position papers: 1. LOINC vs. NPU. 2002. http://www.cumul.ch. Accessed: 2006, out of public access now.Search in Google Scholar
52. Bietenbeck A, Boeker M, Schulz S. NPU, LOINC, and SNOMED CT: a comparison of terminologies for laboratory results reveals individual advantages and a lack of possibilities to encode interpretive comments. J Lab Med 2018;42:267–75.10.1515/labmed-2018-0103Search in Google Scholar
53. Dahlweid F-M, Kämpf M, Leichtle A. Interoperability of laboratory data in Switzerland – a spotlight on Bern. J Lab Med 2018;42:251–8.10.1515/labmed-2018-0072Search in Google Scholar
54. Sabutsch S, Weigl G. Using HL7 CDA and LOINC for standardized laboratory results in the Austrian electronic health record. J Lab Med 2018;42:259–66.10.1515/labmed-2018-0105Search in Google Scholar
55. Aerts J. An alternative CDISC-submission domain for laboratory data (LB) for use with electronic health record data. Eur J Biomed Inform 2015;11:2–9.10.24105/ejbi.2015.11.1.2Search in Google Scholar
56. Food and Drug Administration (FDA). Electronic study data submission, data standards, support for the logical observation identifiers names and codes. Fed Reg 2015;80:27690–1.Search in Google Scholar
57. LOINC Working Group: U.S. Food and Drug Administration (FDA), U.S. National Institutes of Health (NIH), Clinical Data Interchange Standards Consortium (CDISC), and Regenstrief Institute. Recommendations for the Submission of LOINC® Codes in Regulatory Applications to the U.S. Food and Drug Administration. 2017. https://www.fda.gov/media/109376/download. Accessed: 6 Oct 2019.Search in Google Scholar
58. de Haan JB. ELM Cumul Project position papers: 2. Why CUMUL?. 2002. http://www.cumul.ch. Accessed: 2006, out of public access now.Search in Google Scholar
59. McDonald CJ, Schadow G, Suico JG, Heitmann KU. Sprechen Sie LOINC? – Eine Einführung. HL7-Mitteilungen 2000;8:6–11.Search in Google Scholar
60. Semler SC, Neidel P. Labor- und Vitaldatenkommunikation mittels LOINC – Evaluierung eines Standards anhand einer ersten Implementation in Deutschland. Abstracts der 47. Jahrestagung der Deutschen Gesellschaft für Medizinische Informatik, Biometrie und Epidemiologie (GMDS). Informatik, Biometrie und Epidemiologie in Medizin und Biologie 2002;33:357.Search in Google Scholar
61. Duhm-Harbeck P. Gemeinsame Sprache im Labor. UKSH-Hauszeitschrift 2005:3.Search in Google Scholar
62. Michel-Backofen A, Röhrig R, Junger A, Hartmann B, Marquardt K. Abbildbarkeit des strukturierten Vokabulars eines PDMS auf das LOINC Vokabular. In: Klar R, et al. editors. GMDS 50. Jahrestagung 2005, Tagungsband. Bonn: GMDS, 2005:252–5.Search in Google Scholar
63. LOINC User Group. Webseite. www.loinc.de. Accessed: 6 Oct 2019.Search in Google Scholar
64. Müller C, Huesgen G. Datenübertragung zwischen Arztpraxis und Labor: Kommunikation über den 2d-Barcode. Deutsches Ärzteblatt 2002;Suppl. Praxis Computer 1:5–7.Search in Google Scholar
65. Friedrich E, Duhm-Harbeck P. LOINC in Deutschland. HL7-Mitteilungen 2007;22:8–12.Search in Google Scholar
66. Duhm-Harbeck P. Standardisierte Labordatenübermittlung. In: Bruhn HD, Fölsch UR, Schäfer H, editors. LaborMedizin – lndikationen, Methodik und Laborwerte, Pathophysiologie und Klinik, 2nd ed. Stuttgart: Schattauer, 2008:26–30.Search in Google Scholar
67. Pantazoglou E, Thun S. Standardisierte Laboranforderung durch LP-Kodes des Kodiersystems LOINC® zur Steigerung der Interoperabilität. GMDS 2014, 59. Jahrestagung der Deutschen Gesellschaft für Medizinische Informatik, Biometrie und Epidemiologie e.V. (GMDS). Göttingen, German Medical Science GMS Publishing House 2014: DocAbstr. 155.Search in Google Scholar
68. Jakob R. LOINC im medizinischen Laboratorium. J Lab Med 2005;29:6–10.Search in Google Scholar
69. DIMDI. Webseite. www.dimdi.de. Accessed: 6 Oct 2019.Search in Google Scholar
70. Heubeck C, Fiebrig I, Criegee-Rieck M. LOINC Benutzerhandbuch. (Deutsche Übersetzung von: McDonald C, Huff S, Vreeman DJ, Mercer K: LOINC User Guide.) http://www.dimdi.de/dynamic/de/klassi/downloadcenter/loinc-relma/loincrelma/. Accessed: 6 Oct 2019.Search in Google Scholar
71. Zunner C, Bürkle T, Prokosch HU, Ganslandt T. Mapping local laboratory interface terms to LOINC at a German university hospital using RELMA V.5: a semi-automated approach. J Am Med Inform Assoc 2013;20:293–7.10.1136/amiajnl-2012-001063Search in Google Scholar PubMed PubMed Central
72. TMF. Expertengespräch „Terminologien und Ontologien in der medizinischen Forschung und Versorgung“, 29.01.2007, Berlin (TMF/HL7/GMDS) – Anhang 2. In: Rienhoff O, Semler SC, editors. Terminologien und Ordnungssysteme in der Medizin. Berlin: MWV, 2015:207−12.Search in Google Scholar
73. Drepper J, Semler SC. Standardisierung klinischer Forschungsdaten auf Basis von CDISC als Voraussetzung für eine bessere Integration von Forschung und Versorgung. In: Jäckel A, editor. Telemedizinführer Deutschland – Ausgabe 2006. Bad Nauheim, 2005:362–6.Search in Google Scholar
74. Drepper J, Semler SC. Eine gemeinsame Sprache in der Medizin? Datenstandardisierung in medizinischer Versorgung und Forschung. In: Jäckel A, editor. Telemedizinführer Deutschland – Ausgabe 2008. Bad Nauheim, 2007:170–6.Search in Google Scholar
75. BITKOM, VDAP, VHitG, ZVEI. Einführung einer Telematik-Architektur im deutschen Gesundheitswesen – Expertise. Berlin, 2003.Search in Google Scholar
76. IBM Deutschland GmbH, Fraunhofer IAO, SAP Deutschland AG, InterComponentWare AG, ORGA Kartensysteme GmbH. Erarbeitung einer Strategie zur Einführung einer Gesundheitskarte. Standards und Initiativen im Gesundheitswesen – eine evaluierende Übersicht und Empfehlung. Version 1.1. 2004.Search in Google Scholar
77. BearingPoint, Fraunhofer Fokus. eHealth-Planungsstudie Interoperabilität – Ergebnisbericht. Version 2.0. 2014. https://publicwiki-01.fraunhofer.de/Planungsstudie_Interoperabilitaet/index.php/Hauptseite. Accessed: 6 Oct 2019.Search in Google Scholar
78. Semler SC. Standardisierung der elektronischen Befundkommunikation – Nutzen der LOINC-Nomenklatur für die Telematikrahmenarchitektur. In: Jäckel A, editor. Telemedizinführer Deutschland – Ausgabe 2005. Bad Nauheim, 2004:203–8.Search in Google Scholar
79. Rienhoff O, Semler SC, editors. Terminologien und Ordnungssysteme in der Medizin – Standortbestimmung und Handlungsbedarf in den deutschsprachigen Ländern. TMF-Schriftenreihe, Medizinisch Wissenschaftliche Verlagsgesellschaft, Berlin, 2015.10.32745/9783954665174Search in Google Scholar
80. Semler SC, Buckow K, editors. Big Data im deutschen Gesundheitswesen – Handlungsempfehlungen. Eine Bewertung aktueller Möglichkeiten und Herausforderungen. TMF-Schriftenreihe, Medizinisch Wissenschaftliche Verlagsgesellschaft, Berlin, 2019 (in print).Search in Google Scholar
81. Thun S, Heitmann KU, Criegee-Rieck M, Duhm-Harbeck P, Röhrig R, Dugas M, et al. LOINC – eine Sprache die verbindet: Erfahrungsberichte aus verschiedenen Anwendungsbereichen. In: Jäckel A, editor. Telemedizinführer Deutschland – Ausgabe 2009. Bad Nauheim, 2008:305–13.Search in Google Scholar
82. Treinat L. Erste Erfahrungen im Rahmen des Projektes eMeldewesen.nrw – identifizierte Problemlagen und Lösungsansätze. In: Rienhoff O, Semler SC, editors. Terminologien und Ordnungssysteme in der Medizin. Berlin: MWV, 2015:173–80.10.32745/9783954665174-2.12Search in Google Scholar
83. Dugas M, Thun S, Frankewitsch T, Heitmann KU. LOINC® codes for hospital information systems documents: a case study. J Am Med Inform Assoc 2009;16:400–3.10.1197/jamia.M2882Search in Google Scholar
84. Brammen D, Dewenter H, Thiemann V, Majeed RW, Xu T, Heitmann KU, et al. Disseminating a standard for medical records in emergency departments among different software vendors using HL7 CDA. Stud Health Technol Inform 2017;243:132–6.Search in Google Scholar
85. Semler SC, Wissing F, Heyder R. German Medical Informatics Initiative. Methods Inf Med 2018;57:e50–6.10.3414/ME18-03-0003Search in Google Scholar
86. Gehring S, Eulenfeld R. German Medical Informatics Initiative: unlocking data for research and health care. Methods Inf Med 2018;57:e46–9.10.3414/ME18-13-0001Search in Google Scholar
87. Prasser F, Kohlbacher O, Mansmann U, Bauer B, Kuhn KA. Data integration for future medicine (DIFUTURE). Methods Inf Med 2018;57:e57–65.10.3414/ME17-02-0022Search in Google Scholar
88. Haarbrandt B, Schreiweis B, Rey S, Sax U, Scheithauer S, Rienhoff O, et al. HiGHmed – an open platform approach to enhance care and research across institutional boundaries. Methods Inf Med 2018;57:e66–81.10.3414/ME18-02-0002Search in Google Scholar
89. Prokosch H-U, Acker T, Bernarding J, Binder H, Boeker M, Boerries M, et al. MIRACUM: medical informatics in research and care in University medicine. Methods Inf Med 2018; 57:e82–91.10.3414/ME17-02-0025Search in Google Scholar
90. Winter A, Stäubert S, Ammon D, Aiche S, Beyan O, Bischoff V, et al. Smart medical information technology for healthcare (SMITH). Methods Inf Med 2018;57:e92–105.10.3414/ME18-02-0004Search in Google Scholar
91. Ganslandt T, Boeker M, Loebe M, Prasser F, Schepers J, Semler SC, et al. Der Kerndatensatz der Medizininformatik-Initiative: Ein Schritt zur Sekundärnutzung von Versorgungsdaten auf nationaler Ebene [The medical informatics initiative core data set: a step towards the secondary use of routine clinical data on a national scale]. mdi – Forum der Medizin, Dokumentation und Medizin-Informatik 2017;20:17–21.Search in Google Scholar
92. Vreeman DJ, Finnell JT, Overhage JM. A rationale for parsimonious laboratory term mapping by frequency. AMIA Annu Symp Proc 2007:771–5.Search in Google Scholar
93. Schmidt CM. Anderson breaks with IBM Watson, raising questions about artificial intelligence in oncology. J Natl Cancer Inst 2017;109:4–5.10.1093/jnci/djx113Search in Google Scholar
94. Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3:160018.10.1038/sdata.2016.18Search in Google Scholar
95. GO FAIR International Support and Coordination Office (GFISCO). FAIR Principles. https://www.go-fair.org/fair-principles/. Accessed: 8 Oct 2019.Search in Google Scholar
96. Busse R, Blümel M, Knieps F, Bärnighausen T. Statutory health insurance in Germany: a health system shaped by 135 years of solidarity, self-governance, and competition. Lancet 2017;390:882–97.10.1016/S0140-6736(17)31280-1Search in Google Scholar
97. Bundesministerium für Gesundheit. Das Prinzip der Selbstverwaltung. https://www.bundesgesundheitsministerium.de/gesundheitswesen-selbstverwaltung.html. Accessed: 8 Oct 2019.Search in Google Scholar
98. Schubert I, Swart E. Daten für die Versorgungsforschung. Zugang und Nutzungsmöglichkeiten. Gutachten für das Deutsche Institut für Medizinische Dokumentation und Information. DIMDI, 2014.Search in Google Scholar
99. Schepers J, Semler SC. Große Datenmengen in der medizinischen Forschung – Big Data? In: Müller-Mielitz S, Lux T, editors. E-Health-Ökonomie. Wiesbaden: Gabler, 2016:207–36.Search in Google Scholar
100. Schepers J, Semler SC. Große Datenmengen im Versorgungsmonitoring−Big Data? In: Müller-Mielitz S, Lux T, editors. E-Health-Ökonomie. Wiesbaden: Gabler, 2016:383–407.Search in Google Scholar
101. Wegscheider K, Koch-Gromus U. Die Versorgungsforschung als möglicher Profiteur von Big Data. Bundesgesundheitsblatt – Gesundheitsforschung – Gesundheitsschutz 2015;58: 806–12.10.1007/s00103-015-2183-9Search in Google Scholar PubMed
102. Merino-Martinez R, Norlin L, van Enckevort D, Anton G, Schuffenhauer S, Silander K, et al. Toward global biobank integration by implementation of the minimum information about biobank data sharing (MIABIS 2.0 Core). Biopreserv Biobank 2016;14:298–306.10.1089/bio.2015.0070Search in Google Scholar PubMed
103. Kiehntopf M, Böer K, Goebel JW. Biomaterialbanken – Checkliste zur Qualitätssicherung. Berlin: TMF-Schriftenreihe, Medizinisch Wissenschaftliche Verlagsgesellschaft, 2008.Search in Google Scholar
104. Kiehntopf M, Krawczak M. Biobanking and international interoperability: samples. Hum Genet 2011;130:369–76.10.1007/s00439-011-1068-8Search in Google Scholar PubMed
105. Norlin L, Fransson MN, Eriksson M, Merino-Martinez R, Anderberg M, Kurtovic S, et al. A minimum data set for sharing biobank samples, information, and data: MIABIS. Biopreserv Biobank 2012;10:343–8.10.1089/bio.2012.0003Search in Google Scholar PubMed
106. Jacobs G, Wolf A, Krawczak M, Lieb W. Biobanks in the era of digital medicine. Clin Pharmacol Ther 2018;103:761–2.10.1002/cpt.968Search in Google Scholar PubMed PubMed Central
107. Betsou F, Lehmann S, Ashton G, Barnes M, Benson EE, Coppola D, et al. Standard preanalytical coding for biospecimens: defining the sample PREanalytical code. Cancer Epidemiol Biomarkers Prev 2010;19:1004–11.10.1158/1055-9965.EPI-09-1268Search in Google Scholar PubMed
108. Lehmann S, Guadagni F, Moore H, Ashton G, Barnes M, Benson E, et al. Standard preanalytical coding for biospecimens: review and implementation of the Sample PREanalytical Code (SPREC). Biopreserv Biobank 2012;10:366–74.10.1089/bio.2012.0012Search in Google Scholar PubMed PubMed Central
109. Betsou F, Bilbao R, Case J, Chuaqui R, Clements JA, De Souza Y, et al. Standard PREanalytical Code version 3.0. Biopreserv Biobank 2018. doi: 10.1089/bio.2017.0109.Search in Google Scholar PubMed
110. Ganslandt T, Neumeier M. Digital networks for laboratory data: potentials, barriers and current initiatives. Clin Chem Lab Med 2019;57:336–42.10.1515/cclm-2018-1131Search in Google Scholar PubMed
111. Bodenreider O, Cornet R, Vreeman DJ. Recent Developments in Clinical Terminologies – SNOMED CT, LOINC, and RxNorm. Yearb Med Inform 2018;27:129–39.10.1055/s-0038-1667077Search in Google Scholar PubMed PubMed Central
©2019 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Editorial
- Biobanking – current questions and positions
- Articles
- Broad donor consent for human biobanks in Germany and Europe: a strategy to facilitate cross-border sharing and exchange of human biological materials and related data
- Data protection in biobanks from a practical point of view: what must be taken into account during set-up and operation?
- Do we need a biobank law?
- Standardization in biobanking – between cooperation and competition
- Automation in biobanking from a laboratory medicine perspective
- Relevant criteria for the selection of cryotubes. Experiences from the German National Cohort
- Liquid materials for biomedical research: a highly IT-integrated and automated biobanking solution
- Integrated biobanks facilitate high-quality collection and analysis of liquid biomaterials
- LOINC: Origin, development of and perspectives for medical research and biobanking – 20 years on the way to implementation in Germany
- Biobanks for future medicine
Articles in the same Issue
- Frontmatter
- Editorial
- Biobanking – current questions and positions
- Articles
- Broad donor consent for human biobanks in Germany and Europe: a strategy to facilitate cross-border sharing and exchange of human biological materials and related data
- Data protection in biobanks from a practical point of view: what must be taken into account during set-up and operation?
- Do we need a biobank law?
- Standardization in biobanking – between cooperation and competition
- Automation in biobanking from a laboratory medicine perspective
- Relevant criteria for the selection of cryotubes. Experiences from the German National Cohort
- Liquid materials for biomedical research: a highly IT-integrated and automated biobanking solution
- Integrated biobanks facilitate high-quality collection and analysis of liquid biomaterials
- LOINC: Origin, development of and perspectives for medical research and biobanking – 20 years on the way to implementation in Germany
- Biobanks for future medicine


