Abstract
NADH-quinone oxidoreductase (complex I) is the largest and most complicated enzyme complex of the mitochondrial respiratory chain. It is the entry site into the respiratory chain for most of the reducing equivalents generated during metabolism, coupling electron transfer from NADH to quinone to proton translocation, which in turn drives ATP synthesis. Dysfunction of complex I is associated with neurodegenerative diseases such as Parkinson’s and Alzheimer’s, and it is proposed to be involved in aging. Complex I has one non-covalently bound FMN, eight to 10 iron-sulfur clusters, and protein-associated quinone molecules as electron transport components. Electron paramagnetic resonance (EPR) has previously been the most informative technique, especially in membrane in situ analysis. The structure of complex 1 has now been resolved from a number of species, but the mechanisms by which electron transfer is coupled to transmembrane proton pumping remains unresolved. Ubiquinone-10, the terminal electron acceptor of complex I, is detectable by EPR in its one electron reduced, semiquinone (SQ) state. In the aerobic steady state of respiration the semi-ubiquinone anion has been observed and studied in detail. Two distinct protein-associated fast and slow relaxing, SQ signals have been resolved which were designated SQNf and SQNs. This review covers a five decade personal journey through the field leading to a focus on the unresolved questions of the role of the SQ radicals and their possible part in proton pumping.
Introduction: structure of the components of the aerobic respiratory chain with a focus on complex I
Mitochondrial oxidative phosphorylation is one of the most important cellular processes, efficiently providing ATP to all life activities. It is carried out by five multi-subunit transmembrane complexes, designated as complexes I, II, III, IV and V (Figure 1). The first four complexes are electron transfer systems, with complexes I, III and IV generating a proton electrochemical potential, ∆μH+, that drives ATP synthesis through complex V. The initial isolation of complex I from bovine heart mitochondria was reported in 1962 (Hatefi et al., 1962). The subunit composition of the bovine heart complex I including their amino acid sequences was established by John Walker and his colleagues. They also identified all of the core subunits, including all the iron-sulfur proteins (Carroll et al., 2006). It is the largest, with 45 subunits adding to a molecular mass of about 980 kDa, and the most complicated and least understood energy-transducing complex in the aerobic respiratory chain. Bacterial complex I is about 530 kDa and generally composed of 14 subunits (Yagi et al., 1998).
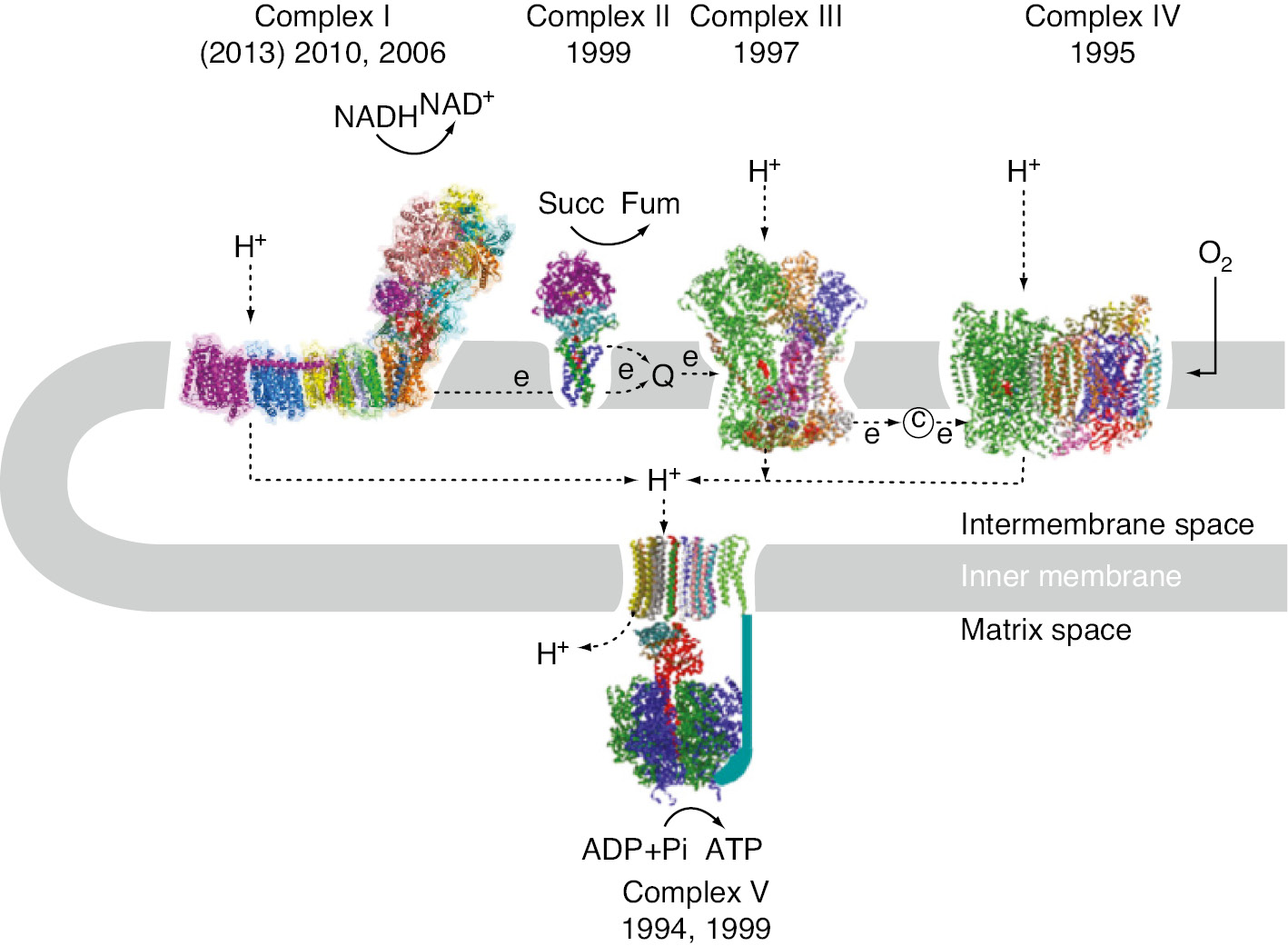
The system of mitochondrial oxidative phosphorylation.
Structural studies on the components of the respiratory chain progressed rapidly after the first report of the X-ray structure of bovine heart F1-component of ATP-synthase in 1994 (Abrahams et al., 1994). X-ray structures of complex IV were reported within 1 year (Iwata et al., 1995; Tsukihara et al., 1995) and structures of complex III (Xia et al., 1997) and complex II (Iverson et al., 1999; Lancaster et al., 1999) were reported within 2-year intervals.
Based on electron microscopy studies, complex I was shown to have a non-symmetric, unique L-shape composed of an extra-membrane and intra-membrane arm (Weiss et al., 1991; Friedrich, 1998; Grigorieff, 1998). The X-ray structure of the extra-membrane part was successfully determined by Sazanov and Hinchliffe at 3.3 Å resolution in a preparation from the thermophilic bacterium Thermus thermophilus HB-8 (Sazanov and Hinchliffe, 2006), followed by the determination of the structure of the Escherichia coli intra-membrane arm at 3.9 Å and the entire complex from T. thermophilus at 4.9 Å resolution (Efremov et al., 2010). This excellent work was continued by reporting the structure of the E. coli intra-membrane arm at 3.0 Å resolution (Efremov and Sazanov, 2011) and, finally, the long-awaited structure of the whole T. thermophilus complex I at 3.3 Å resolution (Baradaran et al., 2013). The X-ray analysis of mitochondrial complex I structure of the obligatory aerobic yeast Yarrowia lipolytica was later reported at 3.6–3.9 Å resolution by Hunte and Brandt’s group (Zickermann et al., 2015). As already deduced from biochemical data, the extra-membrane arm catalyzes the electron transfer reaction while the intra-membrane arm provides the pathways essential for proton translocation. Recently, the structures of the bovine (Vinothkumar et al., 2014; Zhu et al., 2016) and ovine mitochondrial complex I (Fiedorczuk et al., 2016) were determined by means of cryo-electron microscopy at 4.2 and 3.9 Å, respectively. The data revealed that both the electron transfer chain within the extra-membrane arm and the proton pathways of the intra-membrane arm are very similar in bacterial and mitochondrial complex I, although there are subtle but distinct differences (Zickermann et al., 2015; Fiedorczuk et al., 2016; Zhu et al., 2016).
The catalytic component of both, the ATP-synthase and the electron transfer component of complex I extend into the matrix, while the proton-translocation machineries are embedded within the membrane (Figure 1; Walker, 1992; Stock et al., 1999). In contrast, in complex III and IV parts of the electron transfer path involving the b cytochrome (Mitchell, 1975), and cytochrome a and cytochrome a3-CuB (Iwata et al., 1995; Tsukihara et al., 1995), respectively, are localized within integral membrane proteins.
Complex I is one of the largest transmembrane multi-subunit enzyme assemblies (Brandt, 2006; Hirst, 2013) operating as an electron entry point of the electron transfer chain. It catalyzes the following reaction:
where Q stands for (ubi)quinone and suffices N and P indicate that an H+ is on the negative side or positive side of the membrane. Hereafter, we may use Q or quinone as the abbreviation. It is well established that complex I: (i) generates NAD+ from NADH, enabling continued flux through glycolysis and the tri-carboxylic acid cycle; (ii) reduces the quinone pool, so that the electron transport chain can reduce O2 to 2 H2O at the terminal cytochrome c oxidase; (iii) couples the exergonic redox reaction to the vectorial transport of four protons across the mitochondrial inner membrane with a stoichiometry of 4H+/2e− (Wikstrom, 1984; Galkin et al., 1999).
The bacterial and the mitochondrial complex share a set of 14 different subunits that are called the ‘core sub-units’ because they catalyze redox-driven proton translocation. Seven are globular proteins comprising the extra-membrane arm. They contain the NADH oxidation site and the binding sites for all cofactors. The other seven subunits are polytopic membrane proteins that constitute the proton pathways. In addition to these 14 ‘core sub-units’ mitochondrial complex I may contain up to 31 additional, supernumerary subunits (Zickermann et al., 2015; Fiedorczuk et al., 2016; Zhu et al., 2016), that amongst other functions contribute to assembly and regulation of the complex. The positions and identities of all subunits of bovine complex I are now known (Zhu et al., 2015), providing a foundation for an in depth understanding of complex I assembly and mechanism, and its role in cellular metabolism.
The arrangement of redox cofactors in the extra-membrane arm of T. thermophilus HB-8 complex I based on the 3.3 Å structure (Sazanov and Hinchliffe, 2006) is shown in Figure 2. It provides a minimal core model of the historically most well-studied bovine heart complex I. The electron pathway is composed of non-covalently bound FMN (hereafter abbreviated as flavin) and seven different iron-sulfur (abbreviated as Fe/S) clusters. It was revealed as NADH→FMN→N3→N1b→N4→N5→N6a→N6b→N2 arranged as a 95 Å long electron-transfer chain. We have reported a center-to-center distance of 12 Å between the most distal cluster N2 and its direct electron acceptor, the protein-associated ubiquinone species QNf, based on the analysis of their direct spin-spin interaction as described below (Yano et al., 2005). Cluster N7, which is not reducible by NADH, is present only in a limited number of bacteria (Pohl et al., 2007).
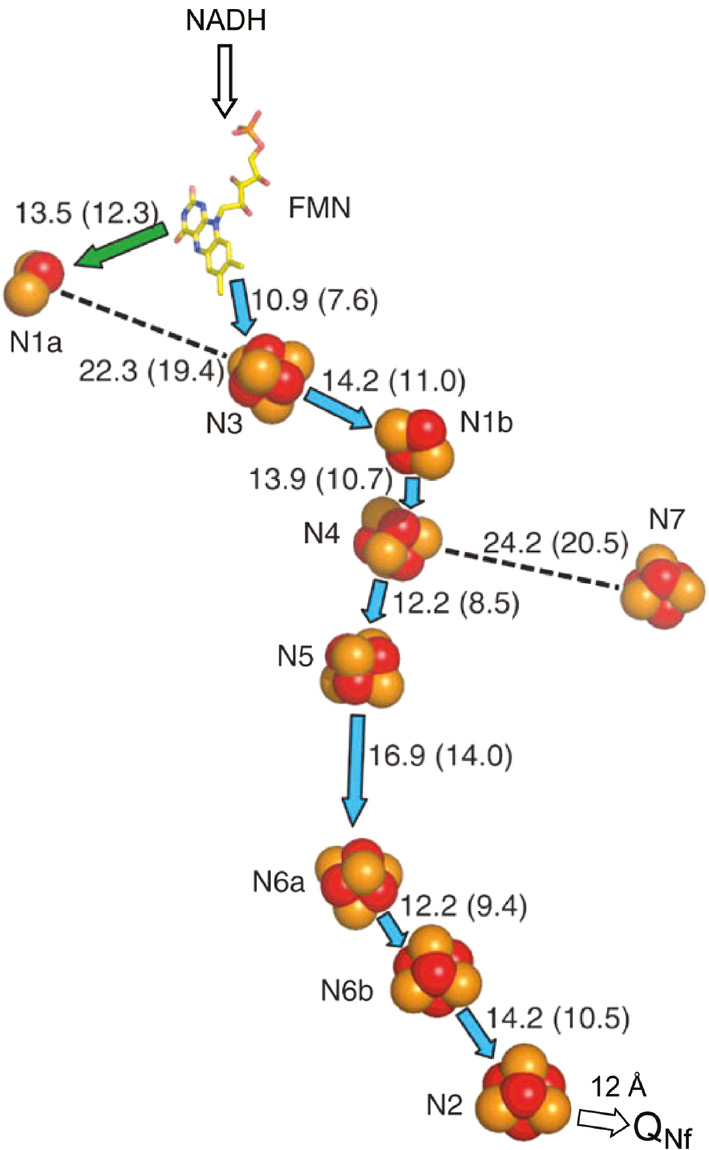
Arrangement of redox cofactors in Thermus thermophilus HB-8 complex I based on the X-ray structure at 3.3 Å resolution (Sazanov and Hinchliffe, 2006).
The center-to-center distance of 12 Å between cluster N2 and protein-associated QNf is based on their spin-spin interaction (Yano et al., 2005).
Because electron paramagnetic resonance (EPR) sample freezing time was at the millisecond level at the time of the early work on complex I, true pre-steady-state reduction kinetics of individual clusters upon the substrate addition was not possible and gave only redox potential information, starting from the reduction of the highest redox midpoint potential (Em7.0) component (Figure 3). Originally, the cluster were named ‘N’ indicating their presence in NADH:ubiquinone oxidoreductase and grouped according to their relaxation behavior as N1, N2, N3 and N4 (Table 1). As more clusters were identified over time, the binuclear clusters were named N1a and N1b and the tetranuclear clusters as N with an ascending suffix (Table 1). Because the two clusters N6 were thought to be tightly coupled they were exceptionally named N6a and N6b (Rasmussen et al., 2001). Throughout the review the nickname of the clusters will be used (Table 1). Cluster N1a has an Em7.0=−370 mV, which is lower than that of the substrate NADH/NAD+ couple (Em7.0=−320 mV), and it is located close to non-covalently bound flavin but away from all other Fe/S clusters. Therefore, it was placed on a side path as a temporary electron deposit site, where it may act as an antioxidant (Sazanov and Hinchliffe, 2006), minimizing the lifetime of flavin semiquinone (SQ) to reduce superoxide O2˙− production (Loschen et al., 1974). All Fe/S clusters from N3 to N6b have an Em7.0=~−250 mV and they were named as ‘isopotential group’ accordingly, while the last cluster N2 has the highest Em7.0 value in the range of −150 to 0 mV (Ingledew and Ohnishi, 1980). The Em7.0 for the Q-pool has been most reliably determined as 90±10 mV for the bacterial reaction center and mitochondria (Dutton et al., 2001). Only N1a and N1b are binuclear [2Fe-2S] clusters; all others are tetranuclear [4Fe-4S] clusters (Palmer, 1985). The X-ray structure of the extra-membranous part of the T. thermophilus complex (Figure 2) confirmed findings from EPR evidence on the number of redox cofactors and their subunit localizations (Ohnishi and Salerno, 1982; Ohnishi, 1998). EPR has been a very informative technique for complex I studies, reviewed in (Ohnishi, 1998). An earlier review of the general characterization of mitochondrial Fe/S clusters by Beinert and Albracht (1982) provides a good background study.
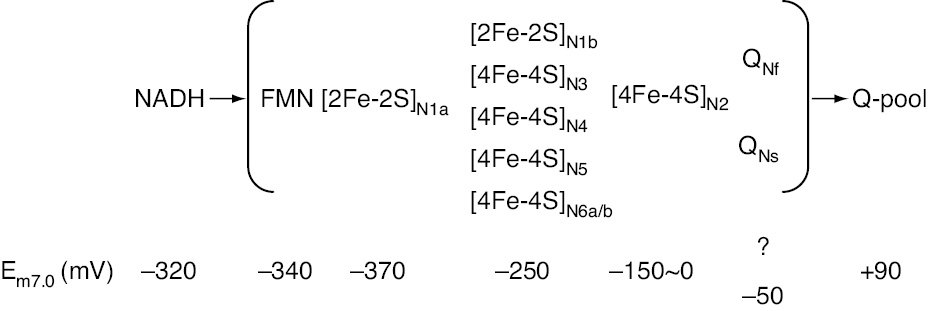
Thermodynamic characterization of redox components in bovine heart complex I.
The iron-sulfur clusters in complex I and their EPR signals.
| Subunit | Fe/S cluster | Nomenclature | Spectral g-values | ||
|---|---|---|---|---|---|
| Nickname | Defined namea | E. coli complex I | Bovine heart complex I | ||
| NuoE/Nqo2 | [2Fe-2S] | N1a | 2Fe[E] | 2.00, 1.95, 1.92 | 2.02, 1.94, 1.92b |
| NuoF/Nqo1 | [4Fe-4S] | N3 | 4Fe[F] | 2.04, 1.92, 1.88 | 2.04, 1.93, 1.86–1.87 |
| NuoG/Nqo3 | [2Fe-2S] | N1b | 2Fe[G] | 2.02, 1.94, 1.94 | 2.02, 1.94, 1.92 |
| [4Fe-4S] | N4 | 4Fe[G]C | 2.09, 1.93, 1.89 | 2.10, 1.94, 1.89 | |
| [4Fe-4S]c | N5 | 4Fe[G]H | 1.90d | 2.07, ~1.93, 1.90 | |
| [4Fe-4S] | N7 | 4Fe[G]e | 2.05, ~1.94, 1.91 | Not present | |
| NuoI/Nqo9 | [4Fe-4S] | N6a | 4Fe[I]1 | 2.09, 1.88, 1.88 | ? |
| [4Fe-4S] | N6b | 4Fe[I]2 | 2.09, 1.94, 1.89 | ? | |
| NuoB/Nqo6 | [4Fe-4S] | N2 | 4Fe[B] | 2.05, 1.91, 1.91 | 2.05, 1.92, 1.92 |
aWe use consensus names as a nickname of Fe/S cluster, and nomenclature proposed by Hirst’s group listed as defined name.
bSomewhat undefined (Em8.0<–500 mV) at extreme low Eh, because of the very low Em of N1a.
cCoordinated by 1His3Cys ligands, all other clusters are 4Cys ligands.
dMost recently, gx=1.90 of cluster N5 was detected with 5 mW and around 3 K in E. coli NuoCDEFG subcomplex. Very high speed helium flow from 4.2 K to ~3 K was assured by Ralph Weber at the EPR division of Bruker BioSpin Corporation (Billerica, MA, USA).
eSignifies that this is a non-conserved form of 4Fe[G]C.
The structure of the bacterial complex I revealed the presence of a more than 110 Å long helix in the intra-membrane arm aligning with the proposed proton pathways (Efremov and Sazanov, 2011). A long surface helix extending parallel to the membrane arm was already described in the electron density map of the Y. lipolytica complex (Hunte et al., 2010). The recent structures obtained by X-ray crystallography and cryo-electron microscopy revealed its presence also in the mitochondrial complex (Zickermann et al., 2015; Fiedorczuk et al., 2016; Zhu et al., 2016). As the overall architecture of F1Fo ATP-synthase was compared to a rotating ‘turbine’, complex I was likened to a ‘steam piston-engine’ with the helix envisioned as a long rod acting as a piston (Efremov et al., 2010; Ohnishi, 2010). However, site-directed mutagenesis of bacterial complex I suggests that this helix is just clamping the membrane arm and providing stability and is not actively involved in catalysis (Belevich et al., 2011; Steimle et al., 2015; Zhu and Vik, 2015). The homologous Na+/H+ antiporter-like subunits, NuoL, M and N (ND5, 4 and 2 in mitochondrial complex I) that provide one putative proton pathway each are arranged in a face-to-back sequence-like nested spoons. It is proposed that a fourth potential proton translocation channel lies at the interface of subunits NuoH, K, J and A (ND1, 4L, 6 and 3 in mitochondrial complex I). The structure indicates that proton translocation in complex I involves coordinated conformational changes in eight symmetrical structural elements. In addition, the structure of the entire complex I from T. thermophilus (Baradaran et al., 2013) shows the presence of linear array of charged amino acids in the intra-membrane arm connecting the putative proton pathways with each other and with the Q-binding site, implying a functional role of these charged residues in proton translocation. The charged residues are mostly lysines rather than carboxylates. These residues were also identified in the mitochondrial complex (Zickermann et al., 2015; Fiedorczuk et al., 2016; Zhu et al., 2016). The proposed Q-binding site is located between the two arms of the complex and is comprised by the globular proteins NuoB and D (PSST and 49 kDa in bovine heart complex I) and the hydrophobic proteins NuoA and H (ND1 and 3, respectively).
As described later, we detected two distinct protein-associated SQ species, designated SQNf and SQNs (SQ molecules in bovine heart NADH-Q oxidoreductase that show a fast and slow relaxation; Magnitsky et al., 2002). In EPR, electron relaxation to the ground state is mediated by energy transfer due to spin transitions coupled to vibrations or phonon transitions with the surrounding lattice. Since the SQNf signal was found to be extremely sensitive to ΔμH+ applied across the mitochondrial inner membrane, most of the EPR analysis was conducted using tightly coupled bovine heart submitochondrial particles (SMP) with a respiratory control ratio (RCR) >8 (Ohnishi et al., 2012; Ohnishi and Ohnishi, 2013). The lack of EPR SQ-signals using the preparation of bacterial complex I might be due to the fact that quinone molecules are completely depleted during complex I purification (Baradaran et al., 2013), limiting the suitability of the T. thermophilus system in the study of protein-associated quinone molecules. Potentiometric titration of flavin and SQNs, both being uncoupler insensitive, was conducted using bovine heart complex I isolated according to Hatefi’s procedure, leading to a preparation which contains 3~4 m of endogenous Q10 per complex I. As will be described later, the Em7.0 values of flavinH2/flavin and QNsH2/QNs are −340 mV and −50 mV, respectively. The Em7.0 value of QNfH2/QNf remains unknown because the determination of QNf SQ requires potentiometric titration in the presence of an applied ΔμH+. This is a technically challenging procedure requiring reconstituted complex I proteoliposomes with RCR>5 capable of maintaining ΔμH+ during the potentiometric titration procedure.
Opening of a new stage of complex I studies
Active and intact mitochondria were first isolated from Saccharomyces cells, as their mitochondrial properties can be modified readily by altering the growth conditions. Saccharomyces carlsbergensis, the yeast utilized for beer production, was employed because it has softer cell walls than Saccharomyces cerevisiae, the yeast used for bread production. An isolation of intact S. carlsbergensis mitochondria that showed a high RCR of >5 upon α-ketoglutarate oxidation and with >3 upon externally added NADH oxidation was reported (Ohnishi et al., 1966a). It was discovered, however, that energy-coupling site I is absent in Saccharomyces, although the second and the third energy-coupling sites are functional as in mammalian mitochondria.
Later, a close similarity of properties of complex I from various species in terms of proton pumping, rotenone sensitivity, and the ‘g=1.94’-type EPR signal from Fe/S clusters was found (Ohnishi et al., 1971; Palmer, 1985). At 77 K, only binuclear iron-sulfur Fe/S clusters such as N1a and N1b are detectable. Around that time, it became obvious that some tetranuclear [4Fe-4S] Fe/S clusters are EPR detectable only below 50 K (G. Palmer, personal communication). Therefore, in order to ascertain that there were no Fe/S clusters from complex I in mitochondria from Saccharomyces, experiments were conducted at temperatures below 77 K (Ohnishi et al., 1971). This led to the unexpected discovery of EPR signals from previously unknown Fe/S clusters in complex I in obligatory aerobic yeasts such as Candida utilis and Endomyces magnusii (Ohnishi et al., 1966b). These Fe/S clusters were not found in Saccharomyces SMPs. Subsequently, bovine heart SMP and isolated bovine heart complex I were examined for the presence of [4Fe-4S] type Fe/S clusters.
EPR spectra of different Fe/S clusters (Figure 4) can be partially resolved by recording spectra at different temperatures and microwave powers. The EPR spectra of Fe/S clusters from slower to faster spin-relaxation rate are observed in the order of cluster N1b→N2→N3→N4. Spectra of N6a and N6b were not recognized at that time because of spectral overlap with N4 and N2 signals, respectively (Sinha et al., 2012). Cluster N5 is EPR detectable only around 3 K at 5 mW microwave power (see Table 1, note d).
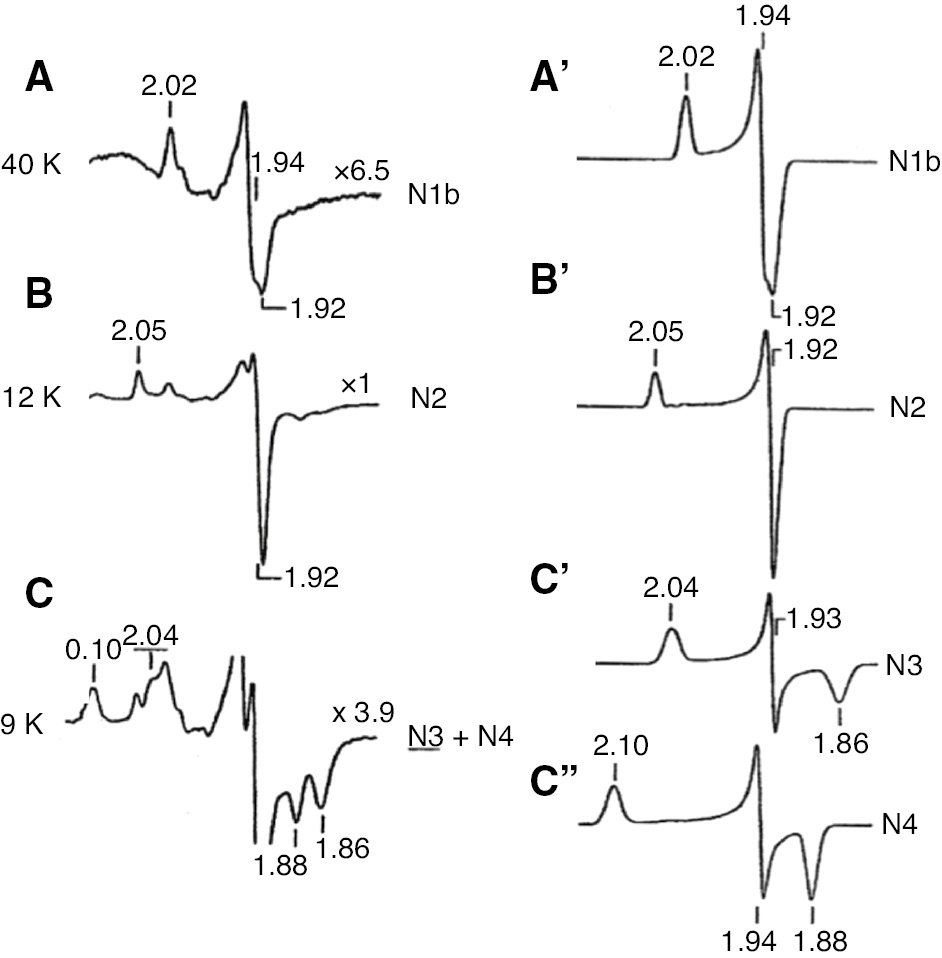
EPR spectra of iron-sulfur clusters in isolated bovine heart complex I reduced by 2 mm NADH.
Spectra were partially resolved at different temperatures. The sample temperatures were: A, 40 K; B, 12 K; C, 9 K. EPR operating conditions: modulation amplitude, 12.5 gauss; microwave power, 5 mW. Computer simulated individual spectra of clusters N1b, N2, N3 and N4 are shown in the right column. Following simulations parameters were used: N1b, gz,y,x=2.02, 1.937, 1.922, Lz,y,x=0.88, 0.81, 1.14 (line width in mT); N2, gz,y,x=2.05, 1.924, 1.917, Lz,y,x=0.84, 0.94, 0.88; N3, gz,y,x=2.034, 1.926, 1.858, Lz,y,x=1.50, 0.70, 1.70; N4, gz,y,x=4.099, 1.936, 1.881, Lz,y,x=1.45, 0.80, 1.20. Reproduced with kind permission from Ohnishi (1998).
Besides studies on mitochondrial complex I, the first author contributed to the discovery of complex I from several bacterial species as summarized in Sled’ et al. (1993). Since a single subunit NADH dehydrogenase from E. coli was characterized by Young’s group in Australia (Rodgers et al., 1982), a complex I counterpart was believed not to be present in E. coli for a long period. In collaboration with K. Matsushita and R. Kaback a complex I counterpart was found in E. coli membranes and named NADH dh I and the single subunit dehydrogenase NADH dh II (Matsushita et al., 1987; Meinhardt et al., 1989). Oshima, the godfather of thermophilic bacteria in Japan, isolated a rotenone sensitive thermophilic bacterium for complex I studies. He named it T. thermophilus HB-8 (HB-8 is the name of the spa from which the strain was isolated). Thermus thermophilus cytoplasmic membranes had Fe/S clusters with much lower Em7.0 values, and a considerably different EPR spectral profile from that of the bovine heart complex I (Meinhardt et al., 1990). This strain endogenously contains solely menaquinone. The lower Em7.0 values of the Fe/S clusters are in accordance with the lower Em7.0 (MQ/MQH2)=−100 mV compared to Em7.0 (UQ/UQH2)= 90 mV. In other collaborations, the first author’s laboratory also studied complex I in Paracoccus denitrificans and Rhodobacter sphaeroides, using a membrane in situ system (Sled’ et al., 1993). Both showed very similar EPR spectral profiles to that of bovine heart complex I.
Protein-associated quinone reactive sites and the role of semiquinone intermediates
An excellent review by Zhu and Gunner (2005) described protein-associated quinones that are found in many transmembrane proteins, coupling electron and proton transfer reactions, such as photosynthetic reaction centers (Okamura et al., 2000; Wraight, 2004), photosystem I (Fromme et al., 2001), ubiquinol-cytochrome c oxidoreductase (complex III; Mitchell, 1975; Lange and Hunte, 2002), and E. coli quinol bo3 oxidase (Yap et al., 2010). There are three major types of quinone-binding systems involving reduction (acceptor), oxidation (donor) and pair-splitting (Murray et al., 1999).
At sites of ‘quinone reduction’ associated with primary dehydrogenases and photosystems, the two-electron reduction of quinone to quinol usually occurs in two sequential one-electron steps, with both electrons supplied by the same (n=1) donor. These sites stabilize the SQ state relative to the quinone and quinol states, approximately equalizing mid-point redox potentials (Em) of the two one-electron couples, Q/Q˙− and Q˙−/QH2 (in general, pKa of QH˙ and QH2 are around 5 and 11, respectively). The stability constant (KStab. 7.0) of free ubiSQ in a hydrophobic milieu (as in the case of Q-pool) was estimated to be approximately 10−10 (Mitchell, 1976). Therefore, the binding of SQ must be at least a factor of ≈105 times tighter than the other two redox states in order to achieve a stability constant of unity. The tight binding of the SQ also allows the site to confine the reactive SQ species, conferring both kinetic and thermodynamic stabilization and preventing unproductive side reactions. In nature, stability constants KStab. of 10–10−2 are seen for efficient converters between 1 and 2 electron transfer processes (Ohnishi et al., 2000). The QN site also stabilizes SQ, and functions as an reduction site (see below). Complex III facilitates reduction of quinone at the Qi site by cytochrome bH by sequential one-electron transfer-steps (Salerno et al., 1979; Crofts and Berry, 1998). While quinone reduction sites have some common properties imposed by the requirements for sequential one electron donation, they achieve these in very different ways and are neither homologs nor structurally related variants.
The E. coli cytochrome bo3 ubiquinol oxidase contains donor sites, which catalyze two electron oxidation of ubiquinol-8. Stabilized SQ radicals associated with this oxidase were first observed by Ingledew et al. (1995). High affinity QH and low affinity QL binding sites of this oxidase have been described (Sato-Watanabe et al., 1995). Recently, both QH and QL have been identified and both can make stable SQ states. The QH site is non-replaceable with Q analogs or specific inhibitors while QL is readily exchangeable with the quinone pool (Yap et al., 2010). The E. coli cytochrome bd quinol oxidase (Ingledew et al., 1992) also contains a site of SQ stabilization that allows reduction of the heme prosthetic groups by pool quinol. This site is functionally similar to the cytochrome bo3 site, but is not homologous to it.
Complex III also possesses a quinol oxidation site at Qo which facilitates the oxidation of quinol to quinone in sequential one-electron transfer steps. However, in this case two electrons are donated to two different accepters with widely different Em values, namely the Rieske Fe/S cluster and cytochrome bL. This is a pair splitting site as proposed originally by Wikström and Berden (1972). The spatial separation of the acceptors and their gross thermodynamic non-equivalence gives the site very different properties from the other sites of quinol oxidation. The approximately 300–400 mV separation in the potential of the acceptors removes the requirement for thermodynamic stabilization of the SQ state at this Qo site (Crofts and Berry, 1998). Pair splitting requires access control as well as different one-electron potentials and is now extensively discussed in bioenergetics as ‘electron bifurcation’ (Peters et al., 2016).
Importance of protein-associated semiquinone species in complex I
Normally spin relaxation of an unpaired π electron associated with an aromatic ring in a magnetically isolated environment is very slow. We can detect such g~2.00 free radical signals even at room temperature and they are easily saturated at a low microwave power at cryogenic temperature. However, as seen in (Figure 5A), the SQ spectrum of tightly coupled bovine heart SMP with a RCR of >8 during aerobic NADH oxidation showed an intense SQ signal even at 16 K and 2 mW. In general, cluster N2 remains highly reduced under this thermodynamic-equilibrium condition. The spin-spin interaction between the much faster-relaxing reduced N2 spins greatly enhances the SQNf-spin relaxation.
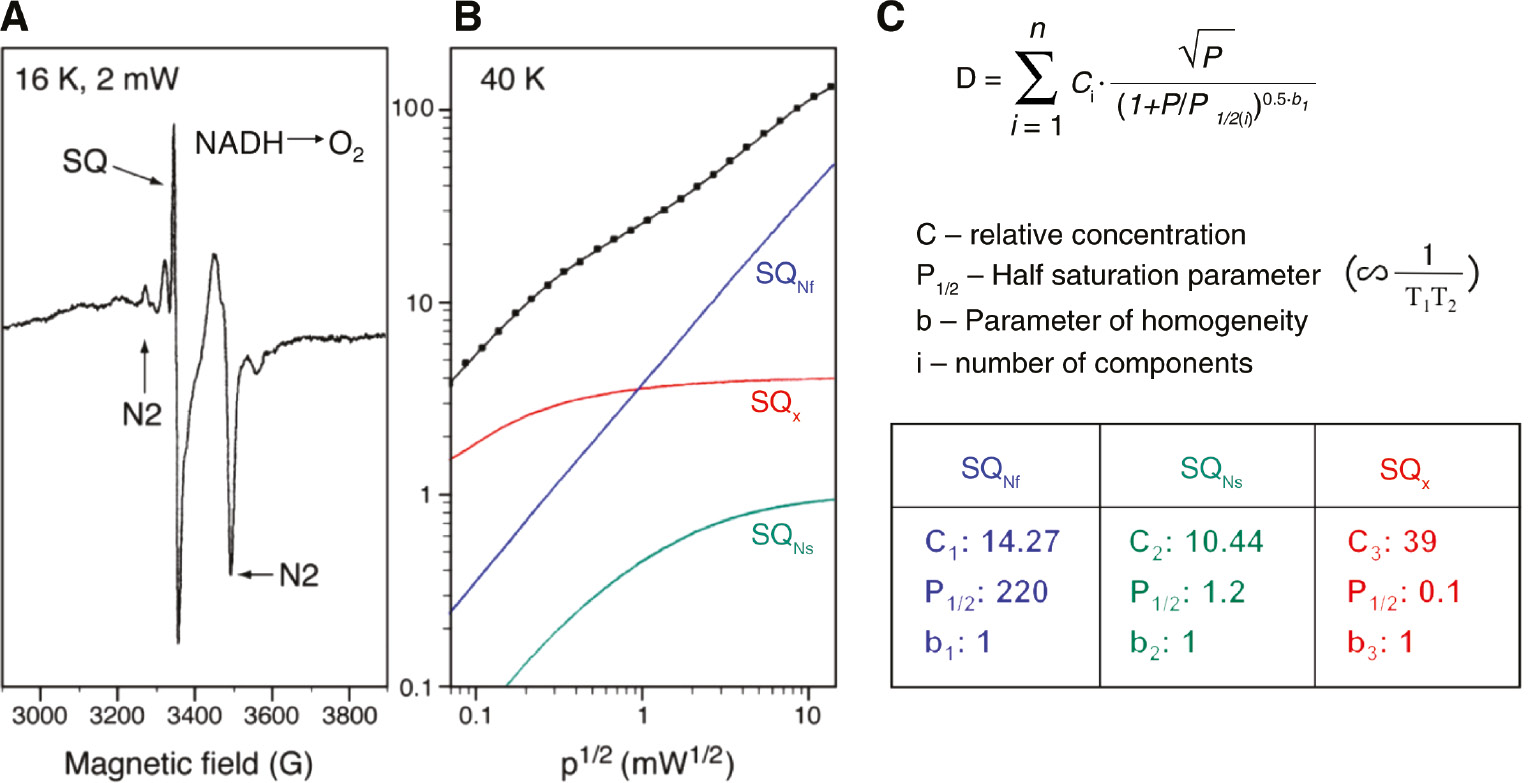
Analysis of power saturation curve from multiple semiquinone species.
(A) EPR spectra of tightly coupled (RCR=8) and activated bovine heart SMP during steady state NADH oxidation. The concave curvature on the right of SQ indicates that CuA2+ of cytochrome c oxidase is in an oxidized state indicating aerobic condition. Sample temperature 16 K, microwave power 2 mW. (B) Power saturation profile of overlapping semiquinone signals. Sample temperature, 40 K. Power saturation data was analyzed by computer fitting to the equation shown in C. (C)
In order to resolve multiple SQ signals, we examined the power saturation profile of the total SQ signal. At 40 K, three different SQ signals were resolved as shown in Figure 5B. Power saturation data can be analyzed by fitting to equation Figure 5C according to Rupp et al. (1978). The tri-phasic power saturation curve was resolved into three individual SQ species, with P1/2 values of 220 (>170) mW, 1.2 mW and 0.1 (<0.1) mW, respectively. We named these three distinct SQ species, SQNf, SQNs and SQX. Both, SQNf and SQNs were quenched by complex I specific inhibitors, such as piericidin A or rotenone, but SQX was not. In addition, SQX species was not found in purified complex I (Ohnishi et al., 2005) and was eliminated mostly by antimycin A and myxothiazol; thus, it must be arising mostly from complex III. The SQNf concentration was <0.3 per complex I and its EPR signal was found to be extremely sensitive to applied ∆μH+ (Ohnishi, 1998; Magnitsky et al., 2002; Yano et al., 2005).
The line shapes of SQNf and SQNs cannot be distinguished in the commonly used X-band (9 GHz) EPR spectrometer. As SQNf is uncoupler sensitive and SQNs is uncoupler insensitive, we could obtain the SQNf spectrum as the difference spectrum in the absence and presence of CCCP at a relatively high microwave power (Figure 6). Tightly coupled bovine heart SMP were prepared routinely with a very low concentration of oligomycin to minimize proton leak through the Fo site of ATP-synthase (Vinogradov, 1998). Prior to reduction with 1.5 mm NADH, SMP were activated with 5 μm NADH (Vinogradov, 1998). The peak to peak line-width (∆Hpp) was ~7.7 gauss, indicating its anionic form (QNf˙−; Bowyer and Ohnishi, 1985).
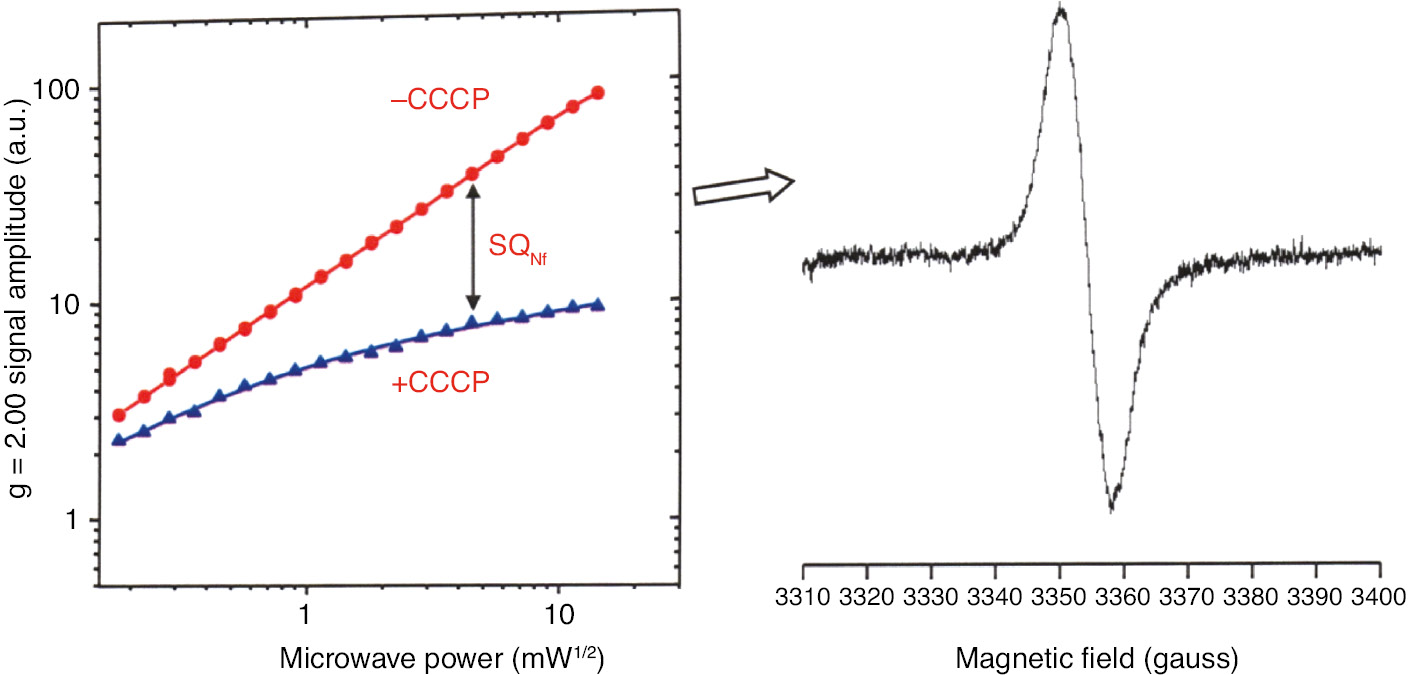
Uncoupler-sensitive SQNf signal can be obtained as the difference spectrum without and with added uncoupler CCCP, at the same microwave power, utilizing tightly coupled bovine heart SMP.
Only SQNs is detected in the presence of CCCP.
The bovine heart complex I preparation from Yoshikawa’s group was reconstituted in proteoliposomes resulting in a RCR>5 when reduced with NADH. With that sample we were able to obtain resolved Q-band (33.9 GHz) EPR spectra of SQNf and SQNs (Figure 7). The principal g values (gzz, gyy, gxx) of SQNf are (2.0045, 2.0036, 2.0005) and of SQNs are (2.0049, 2.0018, 1.9990). Line width (Lz,y,x) is 6.0 gauss for SQNf and 3.5 gauss for SQNs, which differ significantly in spectral resolution from that of X-band EPR. This provided strong supporting experimental evidence for the presence of two distinct protein-associated quinones. Despite these observations, the field has been slow to realize the significance of these observations. Decyl maltoside and dodecyl maltoside are excellent detergents for solubilizing and purifying membrane proteins. These detergents, however, extract most of the lipids and active quinone from complex I, with a simultaneous loss of the sensitivity to complex I specific inhibitors, such as piericidin A and rotenone. However, just adding back lipids alone to complex I restored the inhibitor sensitivity as well as the oxidative phosphorylation capability (Dröse et al., 2002). The essential role of protein-associated quinone molecules was not generally appreciated for a long period, because reconstituted NADH:ubiquinone oxidoreduction did not restore the signatures of native SQ species. The notable exception is the bovine heart complex I isolated by Yoshikawa’s group. Their complex I preparations showed the highest activity and purity obtained to date (Shinzawa-Itoh et al., 2010). This preparation contains one molecule of ubiquinone-10 as cofactor. With this sample, SQNf signals are detectable in the ΔμH+ poised reconstituted complex I proteoliposome system. SQNs EPR signals are also detectable with the isolated and reconstituted complex I without ΔμH+ applied (Shinzawa-Itoh et al., 2010).
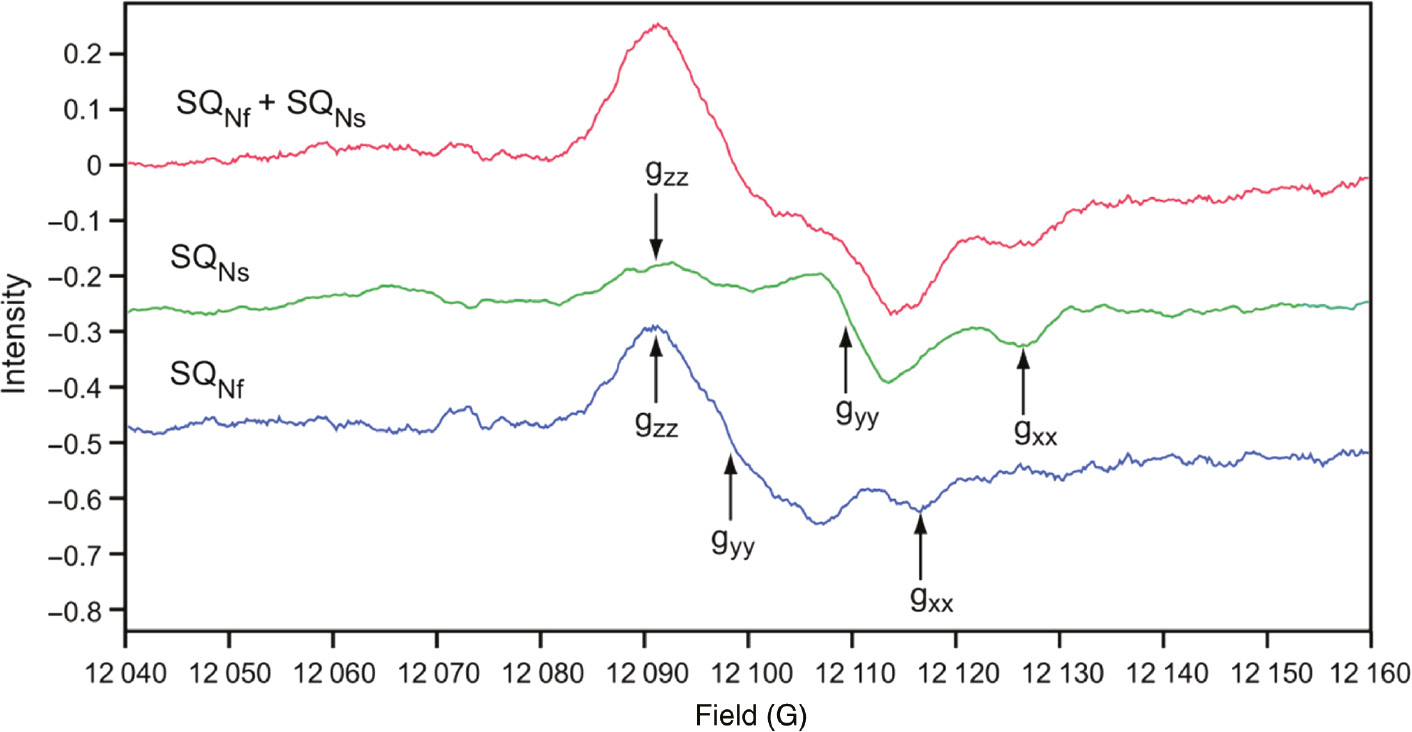
Q-band (33.9 GHz) EPR spectra of SQNf and SQNs: principal g values (gzz, gyy, gxx) for SQNf are (2.0045, 2.0036, 2.0005), and for SQNs (2.0049, 2.0018 and 1.9990).
Line width (Lz,y,x) are 6.0 gauss for SQNf and 3.5 for SQNs. EPR conditions: 150 K; 0.2 mW; modulation 4 gauss; accumulation time 2 h each. Reproduced with kind permission from (Ohnishi et al., 2012).
As reviewed in detail (Ohnishi, 1998; Yano et al., 2005), the 33 gauss splitting of the g//=2.05 signal of cluster N2 (Figure 8A) has been known, but its spin-coupling partner could not be identified, for a long period (Figure 8B). Meticulous temperature dependence analysis of SQNf signal obtained as the difference between steady state NADH-Q1 reductase activity of tightly coupled bovine heart SMP, in the absence and presence of CCCP (Figure 8C) finally gave the optimal EPR condition (Figure 8B) and revealed 56 gauss splitting of the gz,y,x=2.004 of the SQNf spectrum (Yano et al., 2005). The complete Hamiltonian is given in (Figure 9), where the first two terms are electron Zeeman terms which account for the EPR spectra of isolated spin systems, and it involves both the exchange and dipolar interactions (Yano et al., 2005).
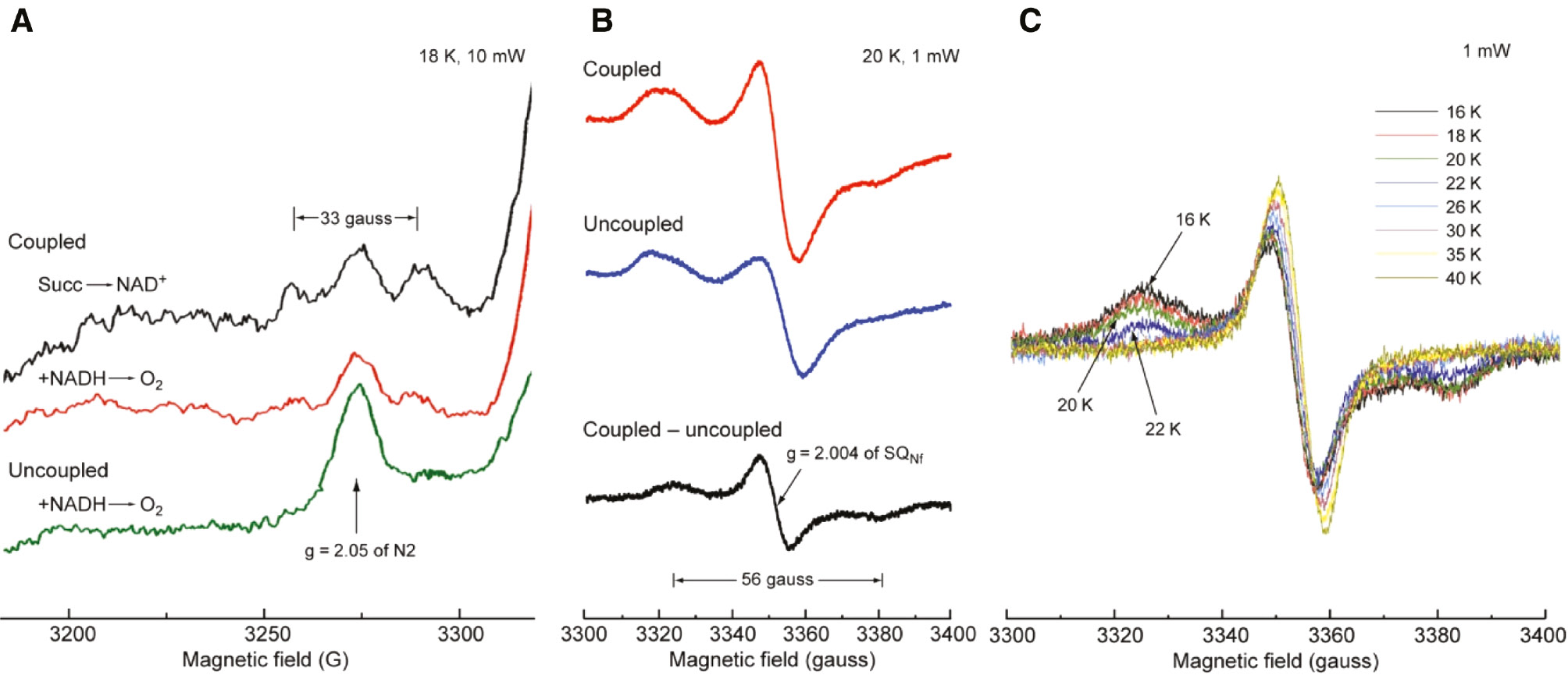
Splitting of the low-field signal of cluster N2.
(A) The green bottom spectrum shows uncoupled NADH reduced state normal single g//=2.05 peak of cluster N2. In coupled state as seen in the middle red, g// peak shows weak 33 gauss splitting. But the aerobic steady state with reversed succinate → NAD+ reaction gives much clearer 33 gauss splitting as seen in the top black spectrum. (B) The 56 gauss splitting (in black) is clearly seen in the g=2.004 SQNf signal as a difference of coupled red spectrum and uncoupled blue spectrum. (C) Detailed temperature dependence of the resolved g=2.004 SQNF signal was shown to select optional Figure (B).
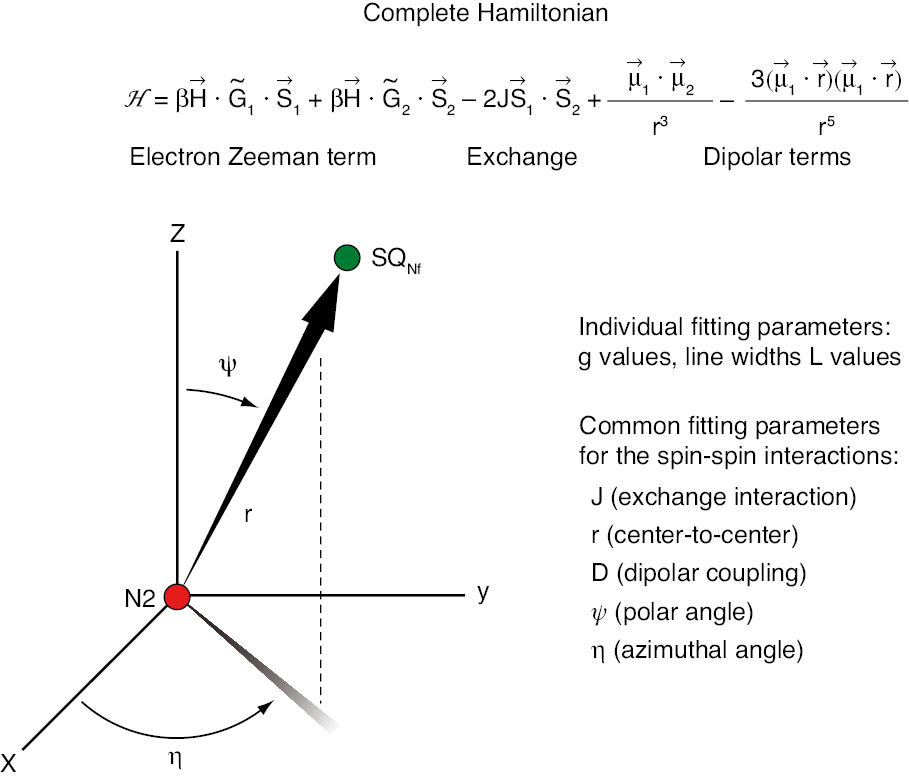
The complete Hamiltonian.
The first two terms are Zeeman terms for EPR spectra of interest including both exchange and dipolar interactions.
The outcome of the simulation of N2-SQNf spin-spin interaction is presented in Figure 10. The center-to-center distance between N2 and SQNf was estimated to be 12 Å, and the projection of the inter-spin vector extends only 5 Å along the membrane normal direction (Yano et al., 2005). This analysis greatly benefitted from the finding that g//=2.05 axis of cluster N2 is perpendicular to the membrane in oriented multilayered preparations (Salerno et al., 1979).
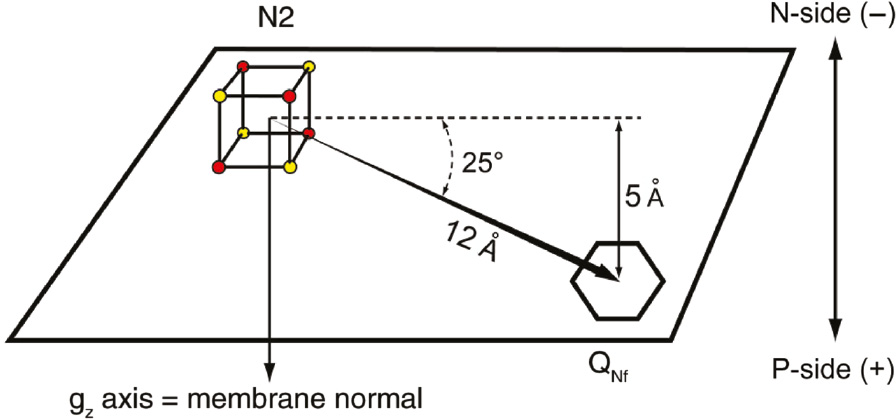
The key outcome of the computer simulation of the spin-spin interaction between cluster N2 and SQNf.
The center-to-center distance between reduced N2 and SQNf is 12 Å. The N2-QNf vector spans only 5 Å of the total ~40 Å thickness of the membrane low dielectric range. Reproduced with kind permission from Ohnishi et al. (2012).
The distance between cluster N2 and QNs was estimated to be about 30 Å, assuming only direct dipolar interaction (Magnitsky et al., 2002). The presence of the second protein-associated SQNs was questioned, primarily because Yoshikawa’s bovine heart complex I contains only one molecule of ubiquinone-10 per complex I. However, it exhibits these two SQ species with identical IC50 for piericidin A but a very different IC50 for rotenone (Magnitsky et al., 2002). It is possible that one quinone alternates between two sites. Relative occupancy may be influenced by factors such as the reduction state of N2, the (de)protonation of nearby residues or local conformational changes.
Proton-pumping mechanism
Based on the results described above, we originally published a hypothesis that the proton-pump of complex I is operated by the redox-driven conformational change of a Q-binding protein, and that the bound form of SQ serves as its gate (Ohnishi and Salerno, 2005). The physical shift between two conformational states changes the thermodynamic stability of QNf, thus gating the proton pump. This pump is connected with the positive side via a proton well, as originally proposed by Mitchell (1976). This hypothesis was different from a reductant-induced oxidation mechanism/reversed Q-cycle model published earlier (Dutton et al., 1998). The framework of the coupling hypothesis was constructed by utilizing EPR techniques before the X-ray structure of complex I became available.
The three largest polytopic subunits ND2, 4, 5/NuoN, M, L (Bos. taurus/E. coli nomenclature) are located at the far end of the membrane arm. They are structurally homologous to each other and to the Mrp-type Na+/H+ antiporters, indicating their involvement in a long distance conformation-coupled indirect proton-pumping mechanism. Ohnishi and colleagues proposed a hypothesis on redox-coupled direct proton-pumping with a 2H+/2e− stoichiometry and simultaneously conformation-coupled indirect proton-pumping for 2H+/2e− located near the NuoL/NuoM (ND4/ND5) subunits (Ohnishi et al., 2010a,c; Figure 11). This hypothesis is based on the finding that QNf˙− is extremely sensitive to ΔμH+. On the other hand, QNs˙− is insensitive to ΔμH+, and binds five orders of magnitude more strongly to the membrane-protein than both fully oxidized (Q) and fully reduced (QH2) states. Therefore, SQNs may function as a converter between one- and two-electron transfer processes (Salerno and Ohnishi, 1980; Crofts and Berry, 1998). According to the model, SQNs also functions as the site of Q/QH2 transfer to the Q-pool. The large conformational change occurs during this redox process, which may induce the opening and closing of ND4 and ND5, homologues of Na+/H+ antiporters (Mathiesen and Hagerhall, 2002). Correspondingly, the third H+/Na+ antiporter-like subunit ND2 acts as the binding site for QNs˙−. We proposed that QNf˙− and QNs˙− sequentially work together as a converter and that these two protein-associated quinone molecules bind to the ND1 and ND2 subunits, respectively (Figure 11; Ohnishi et al., 2010c). This is in good agreement with the fact that these subunits are close to the Q-binding pocket (Zickermann et al., 2015; Fiedorczuk et al., 2016; Zhu et al., 2016). In this model, QNf and QNs work together as converter between n=1 and n=2 electron transfer processes, namely, SQNf-gated direct proton-pump and SQNs-triggered indirect conformation-driven proton pump by the antiporters-like subunits.
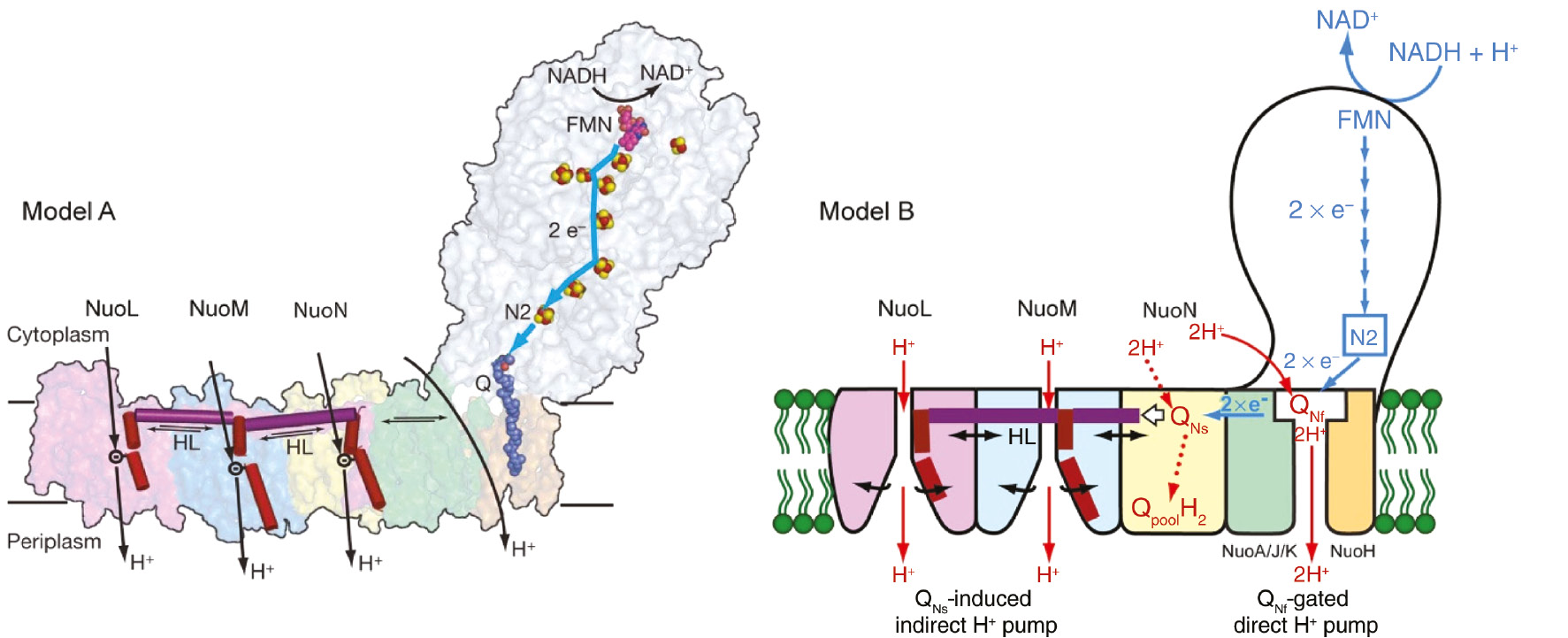
Two hypotheses of proton pumping mechanism of complex I.
Model A: Sazanov’s model which possesses three indirect proton pumping via three antiporter homologs, which creates a 3H+/2e− stoichiometry plus a 1H+/2e− provided by an unknown mechanism. Model B: Ohnishi’s hypothetical model, which involves 2H+/2e− quinone redox coupled direct proton-pump plus 2H+/2e− conformation-coupled indirect proton pump. For QNf-gated direct H+ pump, see explanation in the text.
This model was supported in part by two other findings based on site-directed mutagenesis studies on the ‘rod connected with piston-like’ part of the bacterial and mitochondrial complex I, respectively. Friedrich’s group presented proton translocation by the wild-type complex I and the NuoL-deleted variant after reconstitution into proteoliposomes. A clear lesser quenching of ACMA fluorescence was obtained with liposomes containing the ∆NuoL variant compared to liposomes containing the wild-type (Friedrich, 2001; Steimle et al., 2011). Although the quench of the ACMA fluorescence is proportional to the number of protons pumped in only a small ΔpH range (D’Alessandro et al., 2011), this is a strong indication that the deletion of NuoL decreased the stoichiometry from 4H+/2e− to 2H+/2e−. By deletion of a small accessory subunit of Yarrowia complex I, a stable subcomplex lacking ND4 and ND5 was produced by Brandt’s group and had a 2H+/2e− (see Figure 11; Dröse et al., 2011).
However, other site-directed mutagenesis studies suggested that the horizontal helix is just stabilizing the membrane (Belevich et al., 2011; Steimle et al., 2015; Zhu and Vik, 2015) and the structure of the entire complex I from T. thermophilus (Baradaran et al., 2013) led to a novel model for proton translocation. As seen in Figure 12 the structure of a ‘unique quinone reaction chamber’ was clearly resolved as a 30 Å long and approximately 2−3×4–5 Å wide tube sealed from aqueous medium by hydrophobic elements. As expected, the last electron transfer chain component, Fe/S cluster N2 that directly donates electrons to Q, is located at the end of this chamber.
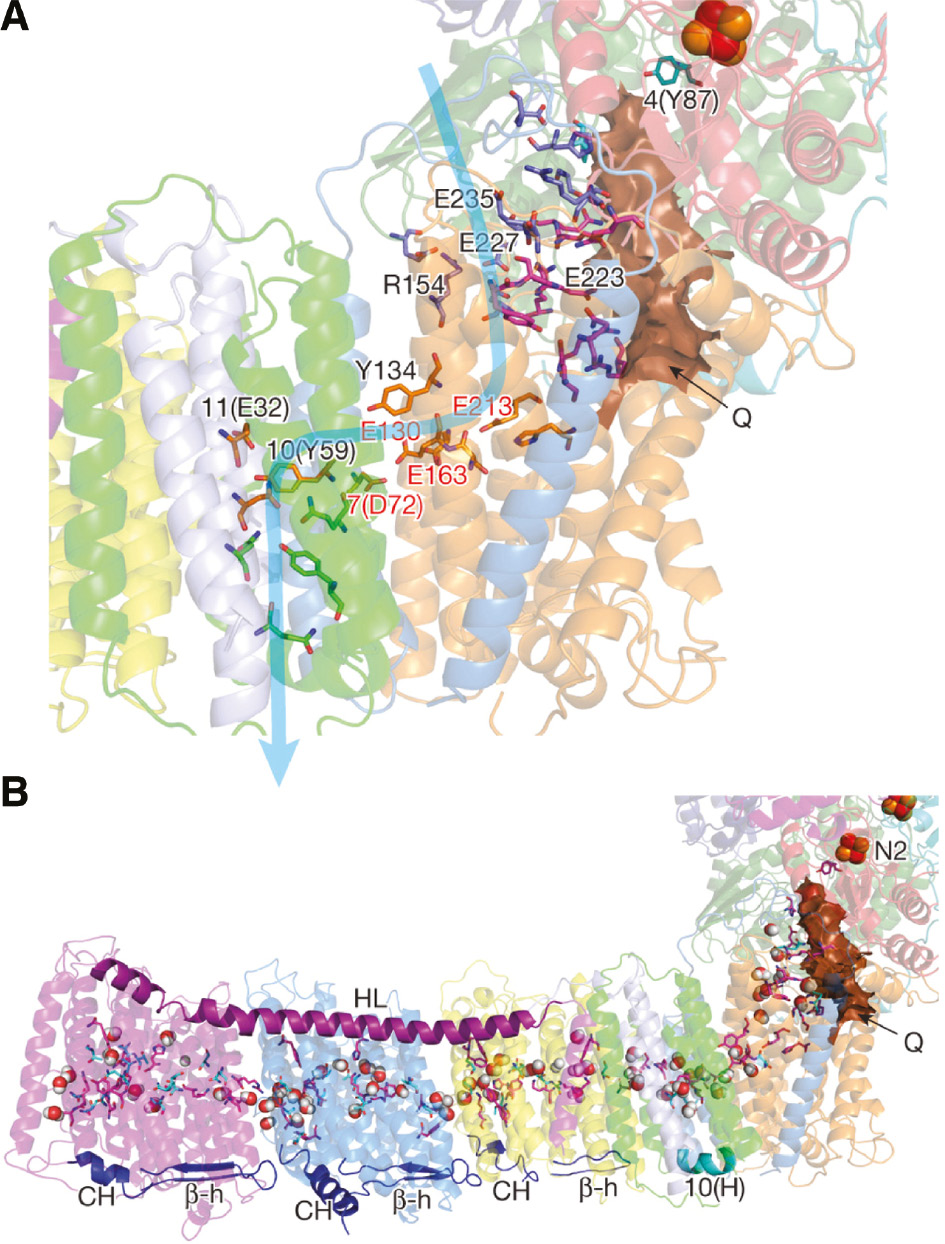
The putative Q-binding site of complex I.
(A) The quinone-reaction chamber in the 3.3 Å resolution structure of T. thermophilus complex I. It appears that the binding site for QNf is 12 Å from N2. (B) Central axis of charged and polar residues. Residues shown are either central to the half-channels of the proton pathways or form the connection between them. Reproduced with kind permission from Baradaran et al. (2013).
In order to determine the position of the quinone-binding site complex I was co-crystallized with piericidin A or the quinone analog decyl-ubiquinone. Both bind in a similar manner, about ~15 Å away from the membrane surface at the deep end of the long narrow cavity. The quinone binding chamber appears to be the binding site for a quinone corresponding to QNf. The distance between N2 and the proposed position of the quinone headgroup of ≤14 Å matches the appropriate distance for efficient electron transfer tunneling (Page et al., 1999). This approximate 12 Å distance is in perfect agreement with that previously determined for QNf by magnetic resonance interaction as described above (Baradaran et al., 2013). The chamber might also accommodate a second pool-derived quinone corresponding to QNs or contain the same quinone in a different environment.
The putative Q entry site (Baradaran et al., 2013) marked with (←Q) in the figure is insulated from solvent and clearly extends well below the lipid bilayer surface. This is approximately 30 Å from the top of the quinone reaction chamber likely corresponding to the SQNs site as previously defined (Ohnishi and Ohnishi, 2013), in agreement with previous distance estimates and its proposed direct connection to the Q-pool. The SQNs↔N2 distance was reported as 30 Å based on the spin relaxation enhancement of SQNs by N2 spins (Magnitsky et al., 2002). The inside of the chamber includes many hydrophilic amino acid residues that could interact with the quinone oxygen atoms (Baradaran et al., 2013). SQNs was proposed to be a converter between n=1 and n=2 electron transfer processes (Ohnishi et al., 2005), which would be facilitated by its location at or near QNf. In a different interpretation of the structural data, the SQNs species was neglected since Yoshikawa’s bovine heart complex I preparation contains only one Q10 molecule (Baradaran et al., 2013). However, both SQ species were detected when this preparation is reconstituted in proteoliposomes (Ohnishi et al., 2005; Yano et al., 2005) either reflecting partition of Q between the two sites or the same quinone in the same site with two different environments. The close correspondence between the structural information deduced from EPR and X-ray analysis is notable, and appears to identify previously observed species with structural sites.
The structure of the entire complex I revealed that NuoH/ND1 adopts a similar structure as the (Na+/H+) antiporter-like subunits and, thus, creates one half-channel, and is connected with the small subunits NuoJ/ND6, NuoA/ND3 and NuoK/ND4L that generate another half-channel (Figure 12). The presence of in total four antiporter homologues correlates with the proposed 4H+/2e− stoichiometry (Baradaran et al., 2013). Additionally, the presence of a fully reduced Q2−-state that remains unprotonated to provide a charge effect to support conformational change of antiporter homologues, was proposed, similar to our earlier hypothesis featuring gating by a charged SQ species (Ohnishi and Salerno, 2005). However, if the inside medium of the quinone reaction chamber is indeed a hydrophilic aqueous environment, the pKa values of the protonation reaction (Q2−→QH−→QH2) would be 13.2 and 11.3, respectively, and protonation would likely occur in spite of various other possibilities as suggested (Baradaran et al., 2013).
Reactive oxygen species, apoptosis, signaling and diseases
Reactive oxygen species (ROS) produced as a side product during cellular respiration are intensively studied because of their role in oxidative stress (Sies et al., 2017). The best known are the superoxide anion, O2˙− (Loschen et al., 1974), hydrogen peroxide, H2O2 (Chance, 1949; Boveris et al., 1972; Chance et al., 1979) and the hydroxyl radical, OH˙ (Ohsawa et al., 2007; Ohta, 2012). The role of H2O2 in eukaryotic cell metabolism started in 1970 (Sies and Chance, 1970). Since then nearly 50 years have passed, yet we still do not have a consensus regarding where O2˙− is produced in complex I. The redox midpoint potential (Em7,0) for an aqueous solution of O2˙−/O2 couple is −0.16 V (Wood, 1988; Petlicki and van de Ven, 1998; Muller, 2000), leading to N2 or SQNf site being suggested as O2˙− generation sites.
We would like to emphasize that the superoxide generation site(s) may vary (Ohnishi et al., 2010b), although it is now generally accepted that ROS formation occurs at the FMN site. In the absence of catalytic electron transfer, superoxide may be generated from flavin SQ (SF). In the presence of electron transfer, for example, with DBQ as an electron acceptor, dynamic shifting of superoxide generation site will occur between SF and protein-associated SQ. When catalytic electron transfer is inhibited by piericidin A or rotenone, superoxide generation site is switched back to SF. Figure 13 shows EPR power saturation profiles of 30 μm bovine heart mitochondrial complex I resulting in a non-saturated Lorentzian-type SF signal with peak-to-peak line-width of about 13 gauss. Figure 13B shows a plateau signal with Gaussian-type protein-associated SQ anion signal with peak-to-peak line width of 7.4 gauss. After an addition of 100 μm DBQ that induces electron transfer, SQ and SF signals are clearly observed (Figure 13C). The addition of rotenone completely eliminates the SQ signal and the spectrum is shifted to the solely SF signal (Figure 13D). The data obtained are summarized in Table 2.
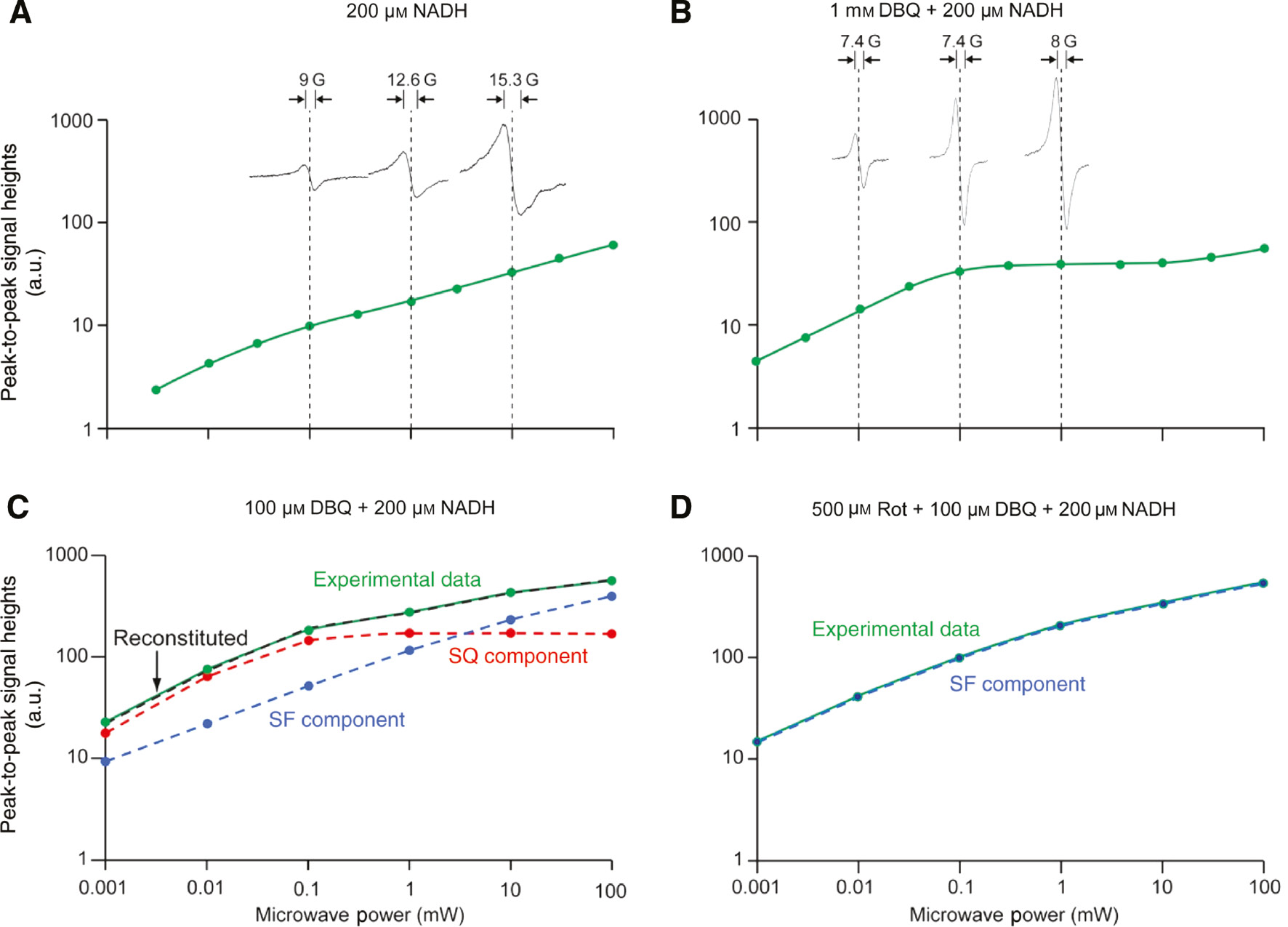
EPR power saturation profiles from 1 to 100 mW of bovine heart complex I.
(A) Bovine heart complex I 30 μm was transferred into an EPR tube and was incubated at 20°C for 3 min. Then, 200 μm NADH was added using the adder-mixer (Ohnishi and Ohnishi, 2010). Sample was frozen in 10 s. (B) After 3 min incubation (at 20°C) of complex I 30 μm with 1 mm DBQ, 200 μm NADH was added and the sample was frozen in 10 s. (C–D) The concentration of complex I in Figures (C–D) was 60 μm, and all tubes were handled similarly. In (C) 100 μm DBQ+200 μm NADH were added to the sample; in (D), 500 μm rotenone+100 μm DBQ+200 μm NADH. In (C), experimental power saturation data (green) was resolved into each component of SF (blue) and SQ (red). The dashed curve (black) in these two figures represents the reconstituted curve, which shows a good fitting to the original experimental data. Other conditions which are common to all these figures: 300 mm sucrose, 40 mm HEPES–HCl (pH 7.8), 0.2% DM and 0.05% DDM. EPR spectra were taken using Bruker EMX spectrometer at 150 K. Reproduced with kind permission from Ohnishi et al. (2010a,b,c).
Shifting between SF and SQ radicals in bovine heart complex I.
| Conditions | SF (%) | SQ (%) |
|---|---|---|
| 0.2 mm NADH | 2.1 | 0.6 |
| 1 mm DBQ+0.2 mm NADH | 1.1 | 6.5 |
| 0.1 mm DBQ+0.2 mm NADH | 1.9 | 2.8 |
| 0.5 mm Rotenone+0.1 mm DBQ+0.2 mm NADH | 3.0 | 0 |
Numbers indicate percentages of both radical forms in the total concentration of complex I (which was assigned as 100).
Acknowledgments
To our great sorrow, T. Ohnishi’s best graduate student and long-term collaborator, John C. Salerno, passed away on December 25, 2015, but he greatly contributed to the biophysical aspects of this review until the end of his life. We would like to dedicate this review article to him. T. Ohnishi would like to express heartfelt gratitude to the previous Department Chairman, Dr. Mark Lemmon for his continued support. He also encouraged his talented assistant, Ms. Patti Thorp, to assist me. Without her great help, I could not have completed this review article. Thanks are also due to Dr. T. Friedrich for his enlightening and stimulating discussions, and to S. Yoshikawa’s group for providing us the most active and pure bovine heart complex I which contains UQ10 as cofactor, but failed to get crystallized, which was provided to our collaborative functional studies. Thanks are also due to Ms. Mary Leonard and to Dr. Akemi Yagi for her professional art and design expertise for all Figures and Tables presented in this review article. I would like to thank Dr. P. Les Dutton and the late Dr. Britton Chance for their support in the earlier years of our research. In closing, I would like to acknowledge the wonderful support from the Perelman School of Medicine at the University of Pennsylvania.
References
Abrahams, J.P., Leslie, A.G., Lutter, R., and Walker, J.E. (1994). Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628.10.1038/370621a0Search in Google Scholar
Baradaran, R., Berrisford, J.M., Minhas, G.S., and Sazanov, L.A. (2013). Crystal structure of the entire respiratory complex I. Nature 494, 443–448.10.1038/nature11871Search in Google Scholar
Beinert, H. and Albracht, S.P. (1982). New insights, ideas and unanswered questions concerning iron-sulfur clusters in mitochondria. Biochim. Biophys. Acta 683, 245–277.10.1016/0304-4173(82)90003-9Search in Google Scholar
Belevich, G., Knuuti, J.,Verkhovsky, M.I., Wikström, M., and Verkhovskaya, M. (2011). Probing the mechanistic role of the long α helix in subunit NuoL in respiratory Complex I in Escherichia coli by site-directed mutagenesis. Mol. Microbiol. 82, 1086–1095.10.1111/j.1365-2958.2011.07883.xSearch in Google Scholar
Boveris, A., Oshino, N., and Chance, B. (1972). The cellular production of hydrogen peroxide. Biochem. J. 128, 617–630.10.1042/bj1280617Search in Google Scholar
Bowyer, J.R. and Ohnishi, T. (1985). EPR spectroscopy in the study of ubisemiquinones in redox chains. In: Coenzyme Q. G. Lenaz, ed. (NY, USA: John Wiley and Sons), pp. 409–432.Search in Google Scholar
Brandt, U. (2006). Energy-converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92.10.1146/annurev.biochem.75.103004.142539Search in Google Scholar
Carroll, J., Fearnley, I.M., Skehel, J.M., Shannon, R.J., Hirst, J., and Walker, J.E. (2006). Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 281, 32724–32727.10.1074/jbc.M607135200Search in Google Scholar
Chance, B. (1949). The enzyme-substrate compounds of horseradish peroxidase and peroxides; kinetics of formation and decomposition of the primary and secondary complexes. Arch. Biochem. 22, 224–252.Search in Google Scholar
Chance, B., Sies, H., and Boveris, A. (1979). Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 59, 527–605.10.1152/physrev.1979.59.3.527Search in Google Scholar
Crofts, A.R. and Berry, E.A. (1998). Structure and function of the cytochrome bc1 complex of mitochondria and photosynthetic bacteria. Curr. Op. Struct. Biol. 8, 501–509.10.1016/S0959-440X(98)80129-2Search in Google Scholar
D’Alessandro, M., Turina, P., and Melandri, B.A. (2011). Quantitative evaluation of the intrinsic uncoupling modulated by ADP and Pi in the reconstituted ATP synthase of Escherichia coli. Biochim. Biophys. Acta 1807, 130–143.10.1016/j.bbabio.2010.08.011Search in Google Scholar PubMed
Dröse, S., Zwicker, K., and Brandt, U. (2002). Full recovery of the NADH:ubiquinone activity of complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica by the addition of phospholipids. Biochim. Biophys. Acta 1556, 65–72.10.1016/S0005-2728(02)00307-9Search in Google Scholar
Dröse, S., Krack, S., Sokolova, L., Zwicker, K., Barth, H.D., Morgner, N., Heide, H., Steger, M., Nubel, E., Zickermann, V., et al. (2011). Functional dissection of the proton pumping modules of mitochondrial complex I. PLoS Biol. 9, e1001128.10.1371/journal.pbio.1001128Search in Google Scholar
Dutton, P.L., Moser, C.C., Sled, V.D., Daldal, F., and Ohnishi, T. (1998). A reductant-induced oxidation mechanism for Complex I. Biochim. Biophys. Acta 1364, 245–257.10.1016/S0005-2728(98)00031-0Search in Google Scholar
Dutton, P.L., Ohnishi, T., Darrouzet, E., Leonard, M.A., Sharp, R.E., Gibney, B.R., Daldal, F., and Moser, C.C. (2001). Coenzyme Q oxidation reduction reactions in mitochondrial electron transport. In: Coenzyme Q: Molecular Mechanisms in Health and Disease. V. E. Kagan and P. J. Quinn, eds. (Boca Raton, USA: CRC Press), pp. 65–82.10.1201/9781420036701.sec1bSearch in Google Scholar
Efremov, R.G. and Sazanov, L.A. (2011). Structure of the membrane domain of respiratory complex I. Nature 476, 414–420.10.1038/nature10330Search in Google Scholar
Efremov, R.G., Baradaran, R., and Sazanov, L.A. (2010). The architecture of respiratory complex I. Nature 465, 441–445.10.1038/nature09066Search in Google Scholar
Fiedorczuk, K., Letts, J.A., Degliesposti, G., Kaszuba, K., Skehel, M., and Sazanov, L.A. (2016). Atomic structure of the entire mammalian mitochondrial complex I. Nature 538, 406–410.10.1038/nature19794Search in Google Scholar
Friedrich, T. (1998). The NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochim. Biophys. Acta 1364, 134–146.10.1016/S0005-2728(98)00024-3Search in Google Scholar
Friedrich, T. (2001). Complex I: a chimaera of a redox and conformation-driven proton pump? J. Bioenerg. Biomembr. 33, 169–177.10.1023/A:1010722717257Search in Google Scholar
Fromme, P., Jordan, P., and Krauss, N. (2001). Structure of photosystem I. Biochim. Biophys. Acta 1507, 5–31.10.1016/S0005-2728(01)00195-5Search in Google Scholar
Galkin, A.S., Grivennikova, V.G., and Vinogradov, A.D. (1999). →H+/2e- stoichiometry in NADH-quinone reductase reactions catalyzed by bovine heart submitochondrial particles. FEBS Lett. 451, 157–161.10.1016/S0014-5793(99)00575-XSearch in Google Scholar
Grigorieff, N. (1998). Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 Å in ice. J. Mol. Biol. 277, 1033–1046.10.1006/jmbi.1998.1668Search in Google Scholar
Hatefi, Y., Haavik, A.G., and Griffiths, D.E. (1962). Studies on the electron transfer system. XL. Preparation and properties of mitochondrial DPNH-coenzyme Q reductase. J. Biol. Chem. 237, 1676–1680.10.1016/S0021-9258(19)83761-4Search in Google Scholar
Hirst, J. (2013). Mitochondrial complex I. Annu. Rev. Biochem. 82, 551–575.10.1146/annurev-biochem-070511-103700Search in Google Scholar PubMed
Hunte, C., Zickermann, V., and Brandt, U. (2010). Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451.10.1126/science.1191046Search in Google Scholar PubMed
Ingledew, W.J. and Ohnishi, T. (1980). An analysis of some thermodynamic properties of iron-sulfur centres in site I of mitochondria. Biochem. J. 186, 111–117.10.1042/bj1860111Search in Google Scholar PubMed PubMed Central
Ingledew, W.J., Rothery, R.A., Gennis, R.B., and Salerno, J.C. (1992). The orientation of the three haems of the ‘in situ’ ubiquinol oxidase, cytochrome bd, of Escherichia coli. Biochem. J. 282, 255–259.10.1042/bj2820255Search in Google Scholar PubMed PubMed Central
Ingledew, W.J., Ohnishi, T., and Salerno, J.C. (1995). Studies on a stabilisation of ubisemiquinone by Escherichia coli quinol oxidase, cytochrome bo. Eur. J. Biochem. 227, 903–908.10.1111/j.1432-1033.1995.tb20217.xSearch in Google Scholar PubMed
Iverson, T.M., Luna-Chavez, C., Cecchini, G., and Rees, D.C. (1999). Structure of the Escherichia coli fumarate reductase respiratory complex. Science 284, 1961–1966.10.1126/science.284.5422.1961Search in Google Scholar PubMed
Iwata, S., Ostermeier, C., Ludwig, B., and Michel, H. (1995). Structure at 2.8-Ångstrom resolution of cytochrome-c-oxidase from Paracoccus denitrificans. Nature 376, 660–669.10.1038/376660a0Search in Google Scholar PubMed
Lancaster, C.R., Kroger, A., Auer, M., and Michel, H. (1999). Structure of fumarate reductase from Wolinella succinogenes at 2.2 Å resolution. Nature 402, 377–385.10.1038/46483Search in Google Scholar PubMed
Lange, C. and Hunte, C. (2002). Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc. Natl. Acad. Sci. USA 99, 2800–2805.10.1073/pnas.052704699Search in Google Scholar
Loschen, G., Azzi, A., Richter, C., and Flohe, L. (1974). Superoxide radicals as precursors of mitochondrial hydrogen peroxide. FEBS Lett. 42, 68–72.10.1016/0014-5793(74)80281-4Search in Google Scholar
Magnitsky, S., Toulokhonova, L., Yano, T., Sled, V.D., Hagerhall, C., Grivennikova, V.G., Burbaev, D.S., Vinogradov, A.D., and Ohnishi, T. (2002). EPR characterization of ubisemiquinones and iron-sulfur cluster N2, central components of the energy coupling in the NADH-ubiquinone oxidoreductase (complex I) in situ. J. Bioenerg. Biomembr. 34, 193–208.10.1023/A:1016083419979Search in Google Scholar
Mathiesen, C. and Hagerhall, C. (2002). Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta 1556, 121–132.10.1016/S0005-2728(02)00343-2Search in Google Scholar
Matsushita, K., Ohnishi, T., and Kaback, H.R. (1987). NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26, 7732–7737.10.1021/bi00398a029Search in Google Scholar
Meinhardt, S.W., Matsushita, K., Kaback, H.R., and Ohnishi, T. (1989). EPR characterization of the iron-sulfur-containing NADH-ubiquinone oxidoreductase of the Escherichia coli aerobic respiratory chain. Biochemistry 28, 2153–2160.10.1021/bi00431a029Search in Google Scholar
Meinhardt, S.W., Wang, D.C., Hon-nami, K., Yagi, T., Oshima, T., and Ohnishi, T. (1990). Studies on the NADH-menaquinone oxidoreductase segment of the respiratory chain in Thermus thermophilus HB-8. J. Biol. Chem. 265, 1360–1368.10.1016/S0021-9258(19)40022-7Search in Google Scholar
Mitchell, P. (1975). The protonmotive Q cycle: a general formulation. FEBS Lett. 59, 137–139.10.1016/0014-5793(75)80359-0Search in Google Scholar
Mitchell, P. (1976). Possible molecular mechanisms of the protonmotive function of cytochrome systems. J. Theoret. Biol. 62, 327–367.10.1016/0022-5193(76)90124-7Search in Google Scholar
Muller, F. (2000). The nature and mechanism of superoxide production by the electron transport chain: its relevance to aging. J. Am. Aging Assoc. 23, 227–253.10.1007/s11357-000-0022-9Search in Google Scholar PubMed PubMed Central
Murray, L., Pires, R.H., Hastings, S.F., and Ingledew, W.J. (1999). Models for structure and function in quinone-binding sites: the Escherichia coli quinol oxidase, cytochrome bo3. Biochem. Soc. Trans. 27, 581–585.10.1042/bst0270581Search in Google Scholar
Ohnishi, T. (1998). Iron-sulfur clusters/semiquinones in complex I. Biochim. Biophys. Acta 1364, 186–206.10.1016/S0005-2728(98)00027-9Search in Google Scholar
Ohnishi, T. (2010). Structural biology: piston drives a proton pump. Nature 465, 428–429.10.1038/465428aSearch in Google Scholar
Ohnishi, S.T. and Ohnishi, T. (2010). An adder-mixer for adding a few microliters of reagent into an electron paramagnetic resonance quartz tube. Anal. Biochem. 406, 89–90.10.1016/j.ab.2010.06.037Search in Google Scholar
Ohnishi, T. and Ohnishi, S.T. (2013). A new trend in the complex I research field. Biol. Chem. 394, 677–683.10.1515/hsz-2013-0117Search in Google Scholar
Ohnishi, T. and Salerno, J.C. (1982). Iron-sulfur clusters in the mitochondrial electron-transport chain. In: Iron-Sulfur Proteins, vol. 4. T. G. Spiro, ed. (NY, USA: Wiley Publishing Co.), pp. 285–327.Search in Google Scholar
Ohnishi, T. and Salerno, J.C. (2005). Conformation-driven and semiquinone-gated proton-pump mechanism in the NADH-ubiquinone oxidoreductase (complex I). FEBS Lett. 579, 4555–4561.10.1016/j.febslet.2005.06.086Search in Google Scholar
Ohnishi, T., Kawaguchi, K., and Hagihara, B. (1966a). Preparation and some properties of yeast mitochondria. J. Biol. Chem. 241, 1797–1806.10.1016/S0021-9258(18)96706-2Search in Google Scholar
Ohnishi, T., Sottocasa, G., and Ernster, L. (1966b). Current approaches to the mechanism of energy-coupling in the respiratory chain. Studies with yeast mitochondria. Bull. Soc. Chim. Biol. (Paris) 48, 1189–1203.Search in Google Scholar
Ohnishi, T., Asakura, T., Yonetani, T., and Chance, B. (1971). Electron paramagnetic resonance studies at temperatures below 77° K on iron-sulfur proteins of yeast and bovine heart submitochondrial particles. J. Biol. Chem. 246, 5960–5964.10.1016/S0021-9258(18)61820-4Search in Google Scholar
Ohnishi, T., Moser, C.C., Page, C.C., Dutton, P.L., and Yano, T. (2000). Simple redox-linked proton-transfer design: new insights from structures of quinol-fumarate reductase. Structure 8, R23–32.10.1016/S0969-2126(00)00098-8Search in Google Scholar
Ohnishi, T., Johnson, J.E. Jr., Yano, T., Lobrutto, R., and Widger, W.R. (2005). Thermodynamic and EPR studies of slowly relaxing ubisemiquinone species in the isolated bovine heart complex I. FEBS Lett. 579, 500–506.10.1016/j.febslet.2004.11.107Search in Google Scholar PubMed
Ohnishi, S.T., Salerno, J.C., and Ohnishi, T. (2010a). Possible roles of two quinone molecules in direct and indirect proton pumps of bovine heart NADH-quinone oxidoreductase (complex I). Biochim. Biophys. Acta 1797, 1891–1893.10.1016/j.bbabio.2010.06.010Search in Google Scholar
Ohnishi, S.T., Shinzawa-Itoh, K., Ohta, K., Yoshikawa, S., and Ohnishi, T. (2010b). New insights into the superoxide generation sites in bovine heart NADH-ubiquinone oxidoreductase (Complex I): the significance of protein-associated ubiquinone and the dynamic shifting of generation sites between semiflavin and semiquinone radicals. Biochim. Biophys. Acta 1797, 1901–1909.10.1016/j.bbabio.2010.05.012Search in Google Scholar
Ohnishi, T., Nakamaru-Ogiso, E., and Ohnishi, S.T. (2010c). A new hypothesis on the simultaneous direct and indirect proton pump mechanisms in NADH-quinone oxidoreductase (complex I). FEBS Lett. 584, 4131–4137.10.1016/j.febslet.2010.08.039Search in Google Scholar
Ohnishi, T., Ohnishi, S.T., Shinzawa-Itoh, K., Yoshikawa, S., and Weber, R.T. (2012). EPR detection of two protein-associated ubiquinone components (SQNf and SQNs) in the membrane in situ and in proteoliposomes of isolated bovine heart complex I. Biochim. Biophys. Acta 1817, 1803–1809.10.1016/j.bbabio.2012.03.032Search in Google Scholar
Ohsawa, I., Ishikawa, M., Takahashi, K., Watanabe, M., Nishimaki, K., Yamagata, K., Katsura, K., Katayama, Y., Asoh, S., and Ohta, S. (2007). Hydrogen acts as a therapeutic antioxidant by selectively reducing cytotoxic oxygen radicals. Nat. Med. 13, 688–694.10.1038/nm1577Search in Google Scholar
Ohta, S. (2012). Molecular hydrogen is a novel antioxidant to efficiently reduce oxidative stress with potential for the improvement of mitochondrial diseases. Biochim. Biophys. Acta 1820, 586–594.10.1016/j.bbagen.2011.05.006Search in Google Scholar
Okamura, M.Y., Paddock, M.L., Graige, M.S., and Feher, G. (2000). Proton and electron transfer in bacterial reaction centers. Biochim. Biophys. Acta 1458, 148–163.10.1016/S0005-2728(00)00065-7Search in Google Scholar
Page, C.C., Moser, C.C., Chen, X., and Dutton, P.L. (1999). Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402, 47–52.10.1038/46972Search in Google Scholar PubMed
Palmer, G. (1985). The electron paramagnetic resonance of metalloproteins. Biochem. Soc. Trans. 13, 548–560.10.1042/bst0130548Search in Google Scholar PubMed
Peters, J.W., Miller, A.F., Jones, A.K., King, P.W., and Adams, M.W. (2016). Electron bifurcation. Curr. Opin. Chem. Biol. 31, 146–152.10.1016/j.cbpa.2016.03.007Search in Google Scholar PubMed
Petlicki, J. and van de Ven, T.G.M. (1998). The equilibrium between the oxidation of hydrogen peroxide by oxygen and the dismutation of peroxyl or superoxide radicals in aqueous solutions in contact with oxygen. J. Chem. Soc. Farad. Transact. 94, 2763–2767.10.1039/a804551hSearch in Google Scholar
Pohl, T., Bauer, T., Dorner, K., Stolpe, S., Sell, P., Zocher, G., and Friedrich, T. (2007). Iron-sulfur cluster N7 of the NADH:ubiquinone oxidoreductase (complex I) is essential for stability but not involved in electron transfer. Biochemistry 46, 6588–6596.10.1021/bi700371cSearch in Google Scholar
Rasmussen, T., Scheide, D., Brors, B., Kintscher, L., Weiss, H., and Friedrich, T. (2001). Identification of two tetranuclear FeS clusters on the ferredoxin-type subunit of NADH:ubiquinone oxidoreductase (complex I). Biochemistry 40, 6124–6131.10.1021/bi0026977Search in Google Scholar
Rodgers, W.H., Springer, W., and Young, F.E. (1982). Cloning and expression of a Streptomyces fradiae neomycin resistance gene in Escherichia coli. Gene 18, 133–141.10.1016/0378-1119(82)90111-1Search in Google Scholar
Rupp, H., Rao, K.K., Hall, D.O., and Cammack, R. (1978). Electron spin relaxation of iron-sulphur proteins studied by microwave power saturation. Biochim. Biophys. Acta 537, 255–260.10.1016/0005-2795(78)90509-3Search in Google Scholar
Salerno, J.C. and Ohnishi, T. (1980). Studies on the stabilized ubisemiquinone species in the succinate-cytochrome c reductase segment of the intact mitochondrial membrane system. Biochem. J. 192, 769–781.10.1042/bj1920769Search in Google Scholar
Salerno, J.C., Blum, H., and Ohnishi, T. (1979). The orientation of iron-sulfur clusters and a spin-coupled ubiquinone pair in the mitochondrial membrane. Biochim. Biophys. Acta 547, 270–281.10.1016/0005-2728(79)90010-0Search in Google Scholar
Sato-Watanabe, M., Itoh, S., Mogi, T., Matsuura, K., Miyoshi, H., and Anraku, Y. (1995). Stabilization of a semiquinone radical at the high-affinity quinone-binding site (QH) of the Escherichia coli bo-type ubiquinol oxidase. FEBS Lett. 374, 265–269.10.1016/0014-5793(95)01125-XSearch in Google Scholar
Sazanov, L.A. and Hinchliffe, P. (2006). Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilius. Science 311, 1430–1436.10.1126/science.1123809Search in Google Scholar PubMed
Shinzawa-Itoh, K., Seiyama, J., Terada, H., Nakatsubo, R., Naoki, K., Nakashima, Y., and Yoshikawa, S. (2010). Bovine heart NADH-ubiquinone oxidoreductase contains one molecule of ubiquinone with ten isoprene units as one of the cofactors. Biochemistry 49, 487–492.10.1021/bi9016318Search in Google Scholar PubMed
Sies, H. and Chance, B. (1970). Then steady-state level of catalase compound I in isolated hemoglobin-free perfused rat liver. FEBS Lett. 11, 172–176.10.1016/0014-5793(70)80521-XSearch in Google Scholar
Sies, H., Berndt, C., and Jones, D.P. (2017). Oxidative stress. Annu. Rev. Biochem. 86, 715–748.10.1146/annurev-biochem-061516-045037Search in Google Scholar
Sinha, P.K., Nakamaru-Ogiso, E., Torres-Bacete, J., Sato, M., Castro-Guerrero, N., Ohnishi, T., Matsuno-Yagi, A., and Yagi, T. (2012). Electron transfer in subunit NuoI (TYKY) of Escherichia coli NADH:quinone oxidoreductase (NDH-1). J. Biol. Chem. 287, 17363–17373.10.1074/jbc.M111.329649Search in Google Scholar
Sled’, V.D., Friedrich, T., Leif, H., Weiss, H., Meinhardt, S.W., Fukumori, Y., Calhoun, M.W., Gennis, R.B., and Ohnishi, T. (1993). Bacterial NADH-quinone oxidoreductases: iron-sulfur clusters and related problems. J. Bioener. Biomembr. 25, 347–356.10.1007/BF00762460Search in Google Scholar
Steimle, S., Bajzath, C., Dörner, K., Schulte, M., Bothe, V., and Friedrich, T. (2011). Role of subunit NuoL for proton translocation by respiratory complex I. Biochemistry 50, 3386–3393.10.1021/bi200264qSearch in Google Scholar
Steimle, S., Schnick, C., Burger, E.M., Nuber, F., Krämer, D., Dawitz, H., Brander, S., Matlosz, B., Schäfer, J., Maurer, K., et al. (2015). Cysteine scanning reveals minor local rearrangements of the horizontal helix of respiratory complex I. Mol. Microbiol. 98, 151–161.10.1111/mmi.13112Search in Google Scholar
Stock, D., Leslie, A.G., and Walker, J.E. (1999). Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705.10.1126/science.286.5445.1700Search in Google Scholar
Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Nakashima, R., Yaono, R., and Yoshikawa, S. (1995). Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science 269, 1069–1074.10.1126/science.7652554Search in Google Scholar
Vinogradov, A.D. (1998). Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim. Biophys. Acta 1364, 169–185.10.1016/S0005-2728(98)00026-7Search in Google Scholar
Vinothkumar, K.R., Zhu, J.P., and Hirst, J. (2014). Architecture of mammalian respiratory complex I. Nature 515, 80–84.10.1038/nature13686Search in Google Scholar PubMed PubMed Central
Walker, J.E. (1992). The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25, 253–324.10.1017/S003358350000425XSearch in Google Scholar
Weiss, H., Friedrich, T., Hofhaus, G., and Preis, D. (1991). The respiratory-chain NADH dehydrogenase (complex I) of mitochondria. Eur. J. Biochem. 197, 563–576.10.1007/978-3-642-77200-9_5Search in Google Scholar
Wikstrom, M. (1984). Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett. 169, 300–304.10.1016/0014-5793(84)80338-5Search in Google Scholar
Wikström, M.K. and Berden, J.A. (1972). Oxidoreduction of cytochrome b in the presence of antimycin. Biochim. Biophys. Acta 283, 403–420.10.1016/0005-2728(72)90258-7Search in Google Scholar
Wood, P.M. (1988). The potential diagram for oxygen at pH 7. Biochem. J. 253, 287–289.10.1042/bj2530287Search in Google Scholar
Wraight, C.A. (2004). Proton and electron transfer in the acceptor quinone complex of photosynthetic reaction centers from Rhodobacter sphaeroides. Front. Biosci. 9, 309–337.10.2741/1236Search in Google Scholar
Xia, D., Yu, C.A., Kim, H., Xia, J.Z., Kachurin, A.M., Zhang, L., Yu, L., and Deisenhofer, J. (1997). Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277, 60–66.10.1126/science.277.5322.60Search in Google Scholar
Yagi, T., Yano, T., Di Bernardo, S., and Matsuno-Yagi, A. (1998). Procaryotic complex I (NDH-1), an overview. Biochim. Biophys. Acta 1364, 125–133.10.1016/S0005-2728(98)00023-1Search in Google Scholar
Yano, T., Dunham, W.R., and Ohnishi, T. (2005). Characterization of the ∆μH+ sensitive ubisemiquinone species (SQNf) and the interaction with cluster N2: new insight into the energy-coupled electron transfer in complex I. Biochemistry 44, 1744–1754.10.1021/bi048132iSearch in Google Scholar PubMed
Yap, L.L., Lin, M.T., Ouyang, H., Samoilova, R.I., Dikanov, S.A., and Gennis, R.B. (2010). The quinone-binding sites of the cytochrome bo3 ubiquinol oxidase from Escherichia coli. Biochim. Biophys. Acta 1797, 1924–1932.10.1016/j.bbabio.2010.04.011Search in Google Scholar PubMed PubMed Central
Zhu, Z. and Gunner, M.R. (2005). Energetics of quinone-dependent electron and proton transfers in Rhodobacter sphaeroides photosynthetic reaction ceters. Biochemistry 44, 82–96.10.1021/bi048348kSearch in Google Scholar PubMed
Zhu, S. and Vik, S. (2015). Constraining the lateral helix of respiratory complex I by cross-linking does not impair enzyme activity or proton translocation. J. Biol. Chem. 290, 20761–20773.10.1074/jbc.M115.660381Search in Google Scholar PubMed PubMed Central
Zhu, J., King, M.S., Yu, M., Klipcan, L., Leslie, A.G., and Hirst, J. (2015). Structure of subcomplex Ibeta of mammalian respiratory complex I leads to new supernumerary subunit assignments. Proc. Natl. Acad. Sci. USA 112, 12087–12092.10.1073/pnas.1510577112Search in Google Scholar PubMed PubMed Central
Zhu, J., Vinothkumar, K.R., and Hirst, J. (2016). Structure of mammalian respiratory complex I. Nature 536, 354–358.10.1038/nature19095Search in Google Scholar PubMed PubMed Central
Zickermann, V., Wirth, C., Nasiri, H., Siegmund, K., Schwalbe, H., Hunte, C., and Brandt, U. (2015). Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 347, 44–49.10.1126/science.1259859Search in Google Scholar PubMed
©2018 Walter de Gruyter GmbH, Berlin/Boston
Articles in the same Issue
- Frontmatter
- Reviews
- Interaction of volatile organic compounds and underlying liver disease: a new paradigm for risk
- Five decades of research on mitochondrial NADH-quinone oxidoreductase (complex I)
- Modifications in small nuclear RNAs and their roles in spliceosome assembly and function
- Minireview
- Transcriptional regulation of human defense peptides: a new direction in infection control
- Research Articles/Short Communications
- Cell Biology and Signaling
- Down-regulated paxillin suppresses cell proliferation and invasion by inhibiting M2 macrophage polarization in colon cancer
- p21Waf1 deficiency does not decrease DNA repair in E1A+cHa-Ras transformed cells by HDI sodium butyrate
- MAC30 knockdown involved in the activation of the Hippo signaling pathway in breast cancer cells
- Inhibition of JAK2/STAT3 signaling suppresses bone marrow stromal cells proliferation and osteogenic differentiation, and impairs bone defect healing
- IL-37 affects the occurrence and development of endometriosis by regulating the biological behavior of endometrial stromal cells through multiple signaling pathways
- Resveratrol alleviates early brain injury following subarachnoid hemorrhage: possible involvement of the AMPK/SIRT1/autophagy signaling pathway
Articles in the same Issue
- Frontmatter
- Reviews
- Interaction of volatile organic compounds and underlying liver disease: a new paradigm for risk
- Five decades of research on mitochondrial NADH-quinone oxidoreductase (complex I)
- Modifications in small nuclear RNAs and their roles in spliceosome assembly and function
- Minireview
- Transcriptional regulation of human defense peptides: a new direction in infection control
- Research Articles/Short Communications
- Cell Biology and Signaling
- Down-regulated paxillin suppresses cell proliferation and invasion by inhibiting M2 macrophage polarization in colon cancer
- p21Waf1 deficiency does not decrease DNA repair in E1A+cHa-Ras transformed cells by HDI sodium butyrate
- MAC30 knockdown involved in the activation of the Hippo signaling pathway in breast cancer cells
- Inhibition of JAK2/STAT3 signaling suppresses bone marrow stromal cells proliferation and osteogenic differentiation, and impairs bone defect healing
- IL-37 affects the occurrence and development of endometriosis by regulating the biological behavior of endometrial stromal cells through multiple signaling pathways
- Resveratrol alleviates early brain injury following subarachnoid hemorrhage: possible involvement of the AMPK/SIRT1/autophagy signaling pathway

