Abstract
Objectives
With the development of genomic technologies, the isolation of genomic DNA (gDNA) from clinical samples has become more important for both clinical diagnostics and research studies. Blood samples collected in PAXgene Blood RNA Tubes, which are typically used for RNA extraction, can be used for gDNA extraction, particularly in clinical studies when only such blood samples are available.
Methods
We optimized the pre-treatment of blood samples collected in PAXgene Blood RNA Tubes. For the first time, three magnetic bead-based automated platforms (QIAsymphony SP, Maxwell RSC and KingFisher Apex) were compared for gDNA extraction from these blood samples. Additionally, the effects of storage at 4 °C or freeze-thaw cycles of the blood samples on gDNA yield were investigated. High-throughput extraction in 96-well format was evaluated.
Results
Systematic optimization of blood sample pre-treatment shows that prolonged incubation at room temperature and/or increased centrifugation speed and time improved gDNA yield from blood samples collected in PAXgene Blood RNA Tubes. QIAsymphony SP (4.27 ± 2.19 µg) and Maxwell RSC (4.82 ± 2.96 µg) produced significantly higher gDNA yields than KingFisher Apex (1.09 ± 0.61 µg). Higher gDNA yields were obtained with shorter storage time at 4 °C or fewer freeze-thaw cycles of the blood samples. In the 96-well format extraction, gDNA yields ranged from 0.24 to 13.46 µg.
Conclusions
Not all magnetic bead-based automated platforms are suitable for gDNA extraction from blood samples collected in PAXgene Blood RNA Tubes. Systematic pre-treatments optimization provides guidance for routine and high-throughput workflows, and storage conditions and freeze-thaw cycles offer practical reference for biobanking.
Introduction
With the development of genomic DNA (gDNA) sequencing techniques, the need for gDNA is increasing in clinical diagnosis and research studies [1]. The gDNA can be extracted from a variety of blood sample types, including blood samples collected in some unconventional tubes [2], [3], [4], [5], [6], [7], [8], [9], [10], [11]. Whole blood collected in PAXgene Blood RNA Tube (referred to as “PAXgene RNA blood samples”) are typically used for RNA extraction. However, when PAXgene RNA blood samples are the only ones available, particularly for clinical studies, they can be used to extract gDNA. Augello et al. [9] developed the PAXgene Blood RNA Tube and evaluated the gDNA extraction from PAXgene RNA blood samples using Qiagen manual kit. Kelly et al. [10] extracted gDNA from 100 µL PAXgene RNA blood samples by using the QIAamp Blood Mini Kit for Short Tandem Repeat (STR) analysis. Kruhoffer et al. [11] used automated platform QIAxtractor with QIAamp DNA Blood Kit from 1 mL PAXgene RNA blood samples; after vacuum concentration, the gDNA samples were analyzed for Single Nucleotide Polymorphisms (SNPs) using GeneChips. All these three groups used silica membrane column-based extraction kits; however, certain procedural details were unclear or inconsistent, which may confuse researchers attempting to extract gDNA from PAXgene RNA Blood samples. Therefore, systematic optimization of sample pre-treatments is essential for obtaining reproducible and high-quality gDNA extraction, not only for silica-based kits but also for platforms based on different techniques.
Silica membrane column-based method and magnetic bead-based method are the two main methods used in commercial DNA extraction kits. To our knowledge, automated platforms for silica membrane column-based extraction have been developed only by Qiagen, whereas most other automated DNA extraction platforms are based on magnetic bead-based technology, including QIAsymphony SP (Qiagen), Maxwell RSC (Promega), KingFisher Apex (Thermo Fisher) and MagNA Pure 24 (Roche). Due to their widespread use and scalability, magnetic bead-based automated platforms provide clear advantages for high-throughput applications.
In this study, we compared three magnetic bead-based automated methods for gDNA extraction. Pre-treatment steps were systematically optimized by investigating the effects of reagents in PAXgene Blood RNA Tube, room temperature incubation time, or centrifugation speed and time. We also observed the impact of freeze-thaw cycles or the storage period of PAXgene RNA blood samples at 4 °C on gDNA yield. Finally, we conducted high-throughput extractions in 96-well format using QIAsymphony SP platform.
Materials and methods
Sample
Peripheral whole blood was collected at NYU Langone Hospital under IRB approval (S16-00122) in 18 PAXgene Blood RNA Tubes (PreAnalytix/Qiagen, Hilden, Germany) and three K2-EDTA Tubes (BD Vacutainer, Becton Dickinson, Franklin Lakes, NJ, USA; hereafter referred to as “EDTA blood”). All samples were de-identified according to the protocol and followed HIPAA regulations. PAXgene Blood RNA samples were stored at −80 °C before extraction.
Pre-treatment of PAXgene RNA blood samples
Since pre-treatment instructions are not provided by the manufacturer, we determined that PAXgene RNA blood samples were thawed and incubated at room temperature for indicated times. After rotating on Tube Revolver Rotator (Thermo Fisher Scientific, Waltham, MA, USA) at speed 10 (about 30 rpm) for 10 min, 1.5 mL of PAXgene RNA blood samples were aliquoted in 1.5 mL EP tubes. A 1.5 mL aliquot of PAXgene RNA blood samples were centrifuged at indicated speed for indicated time, the supernatant was discarded, and the pellet was resuspended with 400 or 450 μL of phosphate buffered saline (PBS, Corning Inc., Corning, NY, USA) by vortex at high speed for 5 min.
DNA extraction
Extract DNA using QIAsymphony SP automated platform
Blood samples or pellet suspensions were extracted using the QIAsymphony SP automated platform (Qiagen, Hilden, Germany) and the commercial DSP DNA Midi Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. After pre-treatment, a 450 μL aliquot was transferred to a new vial for loading onto the QIAsymphony SP and extracted using blood extraction program: DNA_Blood_400_V6_DSP (Qiagen, Hilden, Germany). DNA was eluted with 100 μL of elution buffer.
Among the three magnetic bead-based automated platforms evaluated in this study, two supported high-throughput extraction in 96-well format and one supported 48-well format (Table 1). Considering both gDNA quality and yield, the QIAsymphony SP platform was selected for high-throughput extraction. In high-throughput extraction, the pre-treated blood samples were processed in 96-well format by loading 96 samples onto the QIAsymphony SP system in a single batch.
DNA quality, quantity, and platform characteristics of four DNA extraction methods.
| Extraction method (n≥3) | Kit type | Kit chemistry | Available format | DNA concentration, ng/μL mean ± SD |
A260/A280 mean ± SD |
A260/A230 mean ± SD |
DIN value | Main peakb, kb |
|---|---|---|---|---|---|---|---|---|
| DNeasy blood & tissue kit | Manual | Silica-based | Single tube | 43.06 ± 51.31 | 1.86 ± 0.05 | 3.68 ± 2.38 | 8.53 ± 0.06 | 55.13 ± 6.19 |
| QIAsymphony SP | Automated | Magnetic bead-based | 96-well | 42.73 ± 21.93 | 1.79 ± 0.06 | 1.80 ± 0.85 | 9.40 ± 0.26 | >60 |
| Maxwell RSC | Automated | Magnetic bead-based | 48-well | 48.52 ± 29.57 | 1.85 ± 0.06 | 1.77 ± 0.44 | 7.60 ± 0.66 | 17.24 ± 2.98 |
| KingFisher apex | Automated | Magnetic bead-based | 96-well | 1.36 ± 1.02 | 1.69 ± 0.54 | 1.09 ± 0.61 | N.A. | >60 |
| EDTA controla | 107.04 ± 34.19 | 1.81 ± 0.01 | 2.83 ± 0.09 | 8.57 ± 0.50 | 57.18 ± 3.08 |
-
aEDTA control gDNA was extracted from EDTA blood using the QIAsymphony SP platform. bMain peak corresponds to the highest-intensity DNA band on the TapeStation electropherogram, reported as molecular weight (kb).
Extract DNA using maxwell RSC automated platform
The pellet suspensions were extracted using the Maxwell RSC automated platform (Promega Corporation, Madison, WI, USA) and the commercial Maxwell RSC Whole Blood DNA Kit (Promega Corporation, Madison, WI, USA) following the manufacturer’s instructions. After pre-treatment, a 400 μL of pellet suspension was loaded to the sample well of Maxwell RSC Whole blood DNA Kit and extracted using blood extraction program (Promega Corporation, Madison, WI, USA). DNA was eluted with 100 μL of elution buffer.
Extract DNA using KingFisher apex automated platform
The pellet suspensions were extracted using the KingFisher Apex automated platform (Thermo Fisher Scientific, Waltham, MA, USA) and the commercial MagMAX DNA Multi-Sample Ultra 2.0 Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. After pre-treatment, a 400 μL of pellet suspension was transferred to deep 96-well plate for loading onto the KingFisher Apex and extracted using blood extraction program MagMAX_Ultra2_400 µL_V2 (Thermo Fisher Scientific, Waltham, MA, USA). DNA was eluted with 100 μL of elution buffer.
Extract DNA using Qiagen manual kit
After pre-treatment, a 400 μL of pellet suspension was extracted using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), a silica membrane column-based kit, following the manufacturer’s instructions, and the DNA was eluted with 100 μL of elution buffer.
DNA quantitation
The concentration and the purity (A260/A280 and A260/A230) of DNA were assessed using NanoDrop Microvolume UV–Vis Spectrophotometer (NanoDrop ONEc, Thermo Fisher Scientific, Waltham, MA, USA). DNA integrity was assessed using agarose gel electrophoresis and also checked on the TapeStation 4,200 System (Agilent, Santa Clara, CA, USA) using Genomic DNA Screentape (#5067–5,365) and associated Genomic DNA Reagents (#5067–5,366). The DNA Integrity Number (DIN) value and the highest molecular weight of the main band of DNA was determined using the TapeStation 4,200 software.
Polymerase chain reaction (PCR)
The quality of DNA was assessed by PCR using primers specific to human gDNA, SHGC105883 [12] (Stanford Human Genome Center): forward 5′-GTCAGAAGACTGAAAACGAAGCC-3′ and reverse 5′-GCTTGCCACATCCTTCTTCAAGT-3′, resulting in a PCR product of 295 bp. PCR mixtures contained 12.5 μL of One Taq 2X Master Mix (New England Biolabs Inc., Ipswich, MA, USA), 0.2 μM of each primer (Sigma-Aldrich, St. Louis, MO, USA), 1 μL of DNA (0.95–102 ng) and H2O to a final volume of 25 μL. Samples were amplified as follow: 1 cycle at 94 °C for 30 s; 35 cycles with steps at 94 °C for 30 s, at 58 °C for 30 s, at 68 °C for 1 min; and a final cycle at 68 °C for 5 min on a T100 Thermocycler (Bio-Rad Laboratories, Hercules, CA, USA).
Hematoxylin and eosin staining and imaging
Ethylenediaminetetraacetic acid (EDTA) blood smears or pellet suspension smears from PAXgene RNA blood samples were prepared using standard procedures [13]. The smear slides were stained with H&E [14] and scanned using the Aperio AT2 Whole Slide Scanner (Leica Microsystems, Wetzlar, Germany).
Statistical analysis
All statistics analyses were conducted using GraphPad Prism 10 (GraphPad Software Inc., San Diego, CA, USA). Statistical analyses were performed using appropriate paired or unpaired tests based on data distribution and study design. Normality was assessed using the Shapiro–Wilk test. Parametric tests included paired t-tests and one-way ANOVA; non-parametric tests included Wilcoxon signed-rank test, Friedman test, and Kruskal–Wallis test. A p-value<0.05 was considered statistically significant.
Results
Systematic pre-treatment optimization for gDNA extraction from PAXgene RNA blood samples using the QIAsymphony SP platform
The effect of the reagents in PAXgene blood RNA tube on gDNA extraction
To investigate the effect of reagents in PAXgene Blood RNA Tubes on gDNA extraction, blood samples were thawed and kept at room temperature for 2 h. Then, 450 µL of whole blood was centrifuged at 5000×g for 10 min to collect the pellet. Another 450 µL aliquot of whole blood was centrifuged at 5000×g for 10 min, the pellet was resuspended in 1 mL of PBS, and then centrifuged at 5000×g for 10 min to collect the washed pellet. Both the pellet and the washed pellet were resuspended in 450 µL of PBS, and the suspension was extracted using the QIAsymphony SP platform. These samples were compared to the direct extraction of 450 µL of whole blood from PAXgene Blood RNA Tubes. The gDNA elution from the whole blood samples were brown and could not be measured for concentration by using NanoDrop Microvolume UV–Vis Spectrophotometer. The DNA yield from the pellet suspension was 3.09 ± 2.04 µg (n=3), while the DNA yield from the washed pellet was 2.03 ± 1.25 µg (n=3), with no significant difference between the two groups (paired t-test, p=0.145, Figure 1A). Gel electrophoresis of gDNA (1 μL) revealed clear DNA bands over 25 kb from both the pellet suspension and the washed pellet (Figure 1B). These results indicate that collecting the pellet by centrifugation is necessary, but washing the pellet with PBS is not.
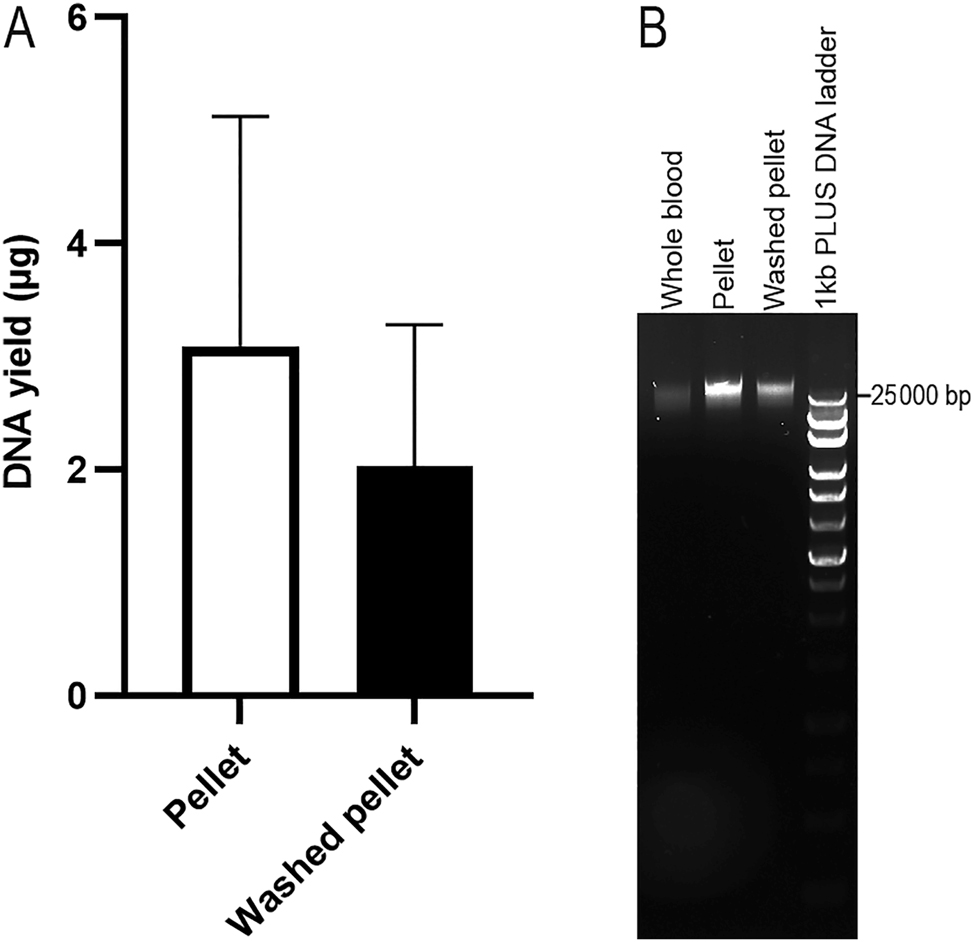
The effect of the reagents in PAXgene blood RNA tube on DNA yield. The gDNA was extracted using the QIAsymphony SP automated platform (n=3, paired samples). DNA yield was assessed using a NanoDrop spectrophotometer (A). Agarose gel electrophoresis of these DNA is shown in (B).
The pellet from the PAXgene RNA blood sample was observed (Figure 2B) to contain many cells with intact, morphologically identifiable nuclei (blue), which are the source of gDNA. Compared to EDTA blood (Figure 2A), the cells from PAXgene RNA blood samples are morphologically much smaller.
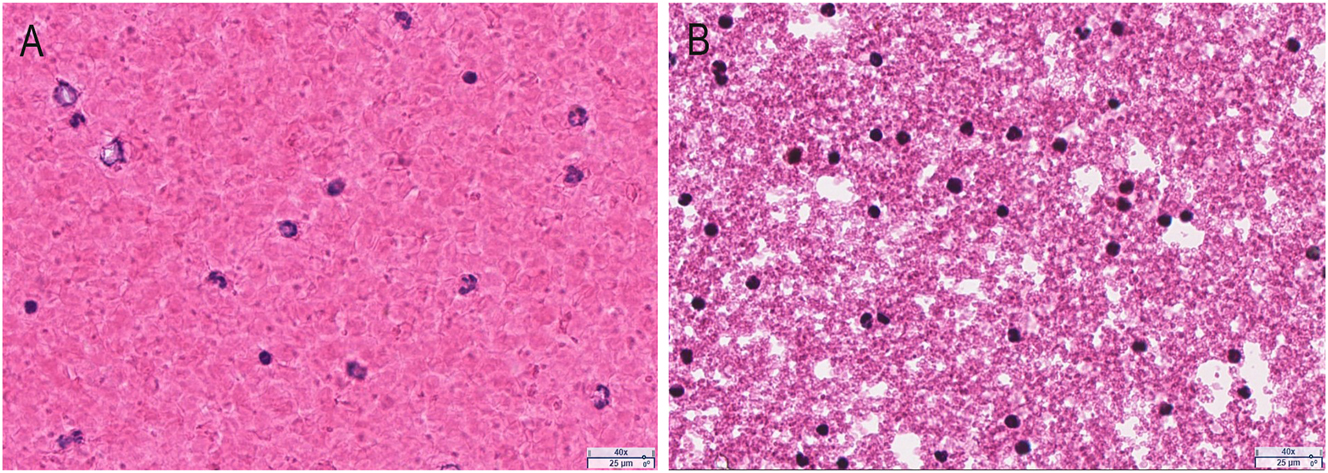
The pellet from PAXgene RNA blood samples (40 × magnification). The smear slides were prepared from EDTA blood (A) and the pellet suspension from PAXgene RNA blood samples (B). After staining with H&E, the slides were scanned using the aperio AT2 whole slide scanner.
Optimization of centrifugation conditions for gDNA extraction from PAXgene RNA blood samples
The gDNA from 400 µL of EDTA blood is sufficient for whole-genome sequencing [5]. Since 1.5 mL of PAXgene RNA blood sample is equivalent to 400 µL of EDTA blood, this volume was used for method development in this study.
The PAXgene RNA blood samples were thawed and kept at room temperature for 2 h. To investigate the effect of centrifugation speed, 1.5 mL blood aliquots were centrifuged at 5000×g [9], 10000×g, or 17000×g for 10 min to collect the pellet. The pellet was resuspended in 450 µL of PBS, and the suspension was extracted using the QIAsymphony SP platform. The DNA yield was 6.27 ± 3.24 μg at 5000×g, 6.90 ± 4.71 μg at 10000×g, and 7.52 ± 4.39 μg at 17000×g (Figure 3A, n=3), with no significant difference (Friedman test, p>0.99). To determine the optimal centrifugation time, 1.5 mL of PAXgene RNA blood samples were centrifuged at 17000×g for 5 min or 10 min. The DNA yield was higher at 10 min (4.85 ± 4.29 µg) compared to 5 min (3.87 ± 2.04 µg) with no significant difference (Figure 3B, n=3, Wilcoxon test, p=0.75).
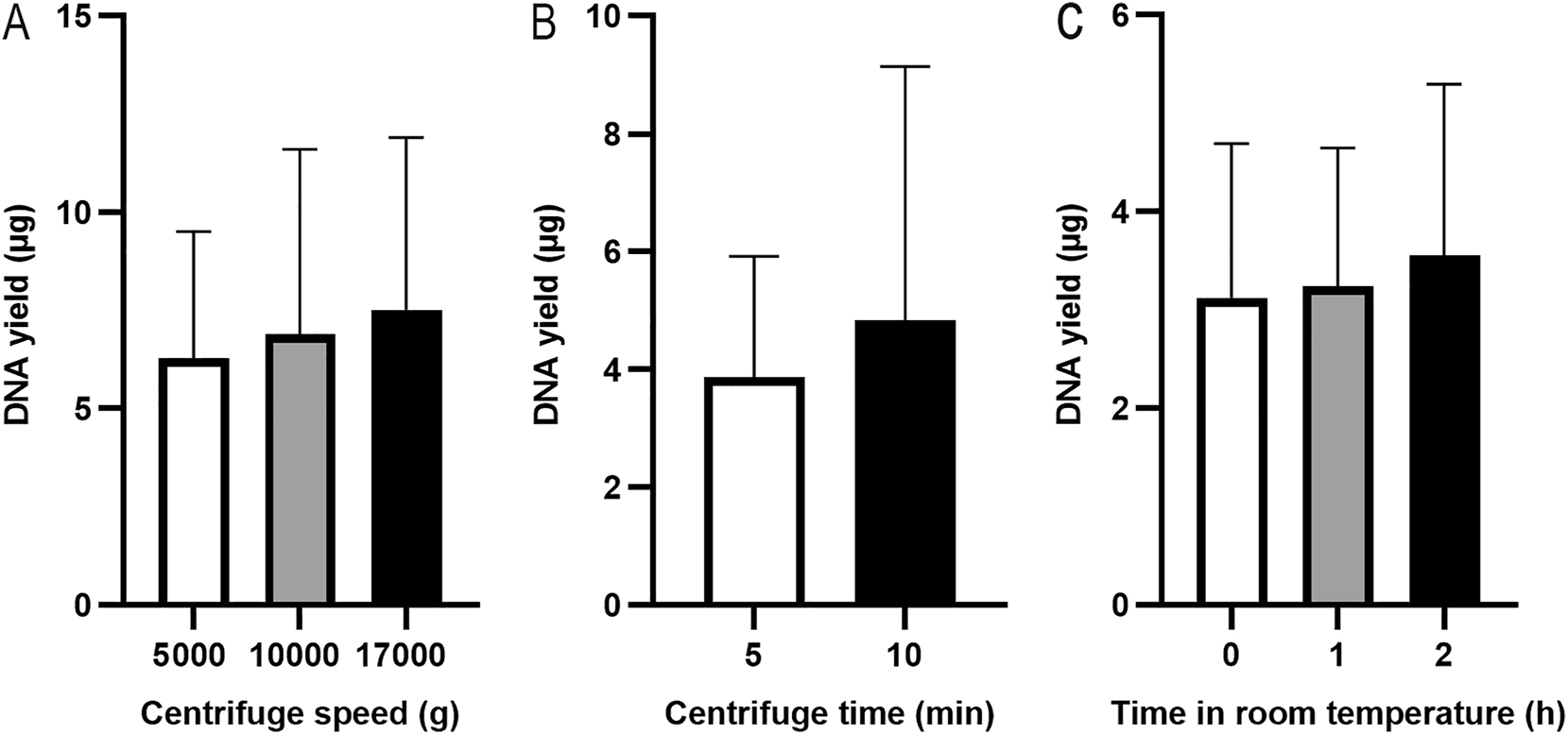
DNA yields from different extraction conditions. Pellets were collected at various centrifuge speeds (A) (n=3, paired samples) and at 17000×g for 5 or 10 min (B) (n=3, paired samples). PAXgene RNA blood samples were thawed and kept at room temperatures for 0, 1 or 2 h (C) (n=3, paired samples). The DNA was extracted using the QIAsymphony SP automated platform, and DNA yield was assessed using a NanoDrop spectrophotometer.
The effect of room temperature incubation on gDNA extraction from PAXgene RNA blood samples
The handbook of PAXgene Blood RNA Kit [15] mentioned that after thawing, incubating PAXgene RNA blood samples at room temperature for 2 h before extraction is crucial for RNA extraction. To determine whether this step is also important for gDNA extraction, the PAXgene RNA blood samples were kept at room temperatures for 0, 1, or 2 h before processing. The 1.5 mL of PAXgene RNA blood samples was centrifuged at 17000×g for 10 min. The pellet was resuspended in 450 µL of PBS, and the suspension was extracted using the QIAsymphony SP platform. The DNA yield was 3.12 ± 1.57 µg after the PAXgene RNA blood samples were thawed at room temperature for 0 h, 3.24 ± 1.40 µg for 1 h, and 3.55 ± 1.74 µg for 2 h (Figure 3C, n=3), with no significant difference in DNA yield among the time points (Friedman test, p=0.36).
gDNA extraction from PAXgene RNA blood samples using four commercial kits
The PAXgene RNA blood samples were thawed and kept at room temperature for 2 h. Pellets were collected from 1.5 mL of blood samples by centrifugation at 17000×g for 10 min. The pellet was resuspended in 400 or 450 µL of PBS, and gDNA was extracted using four different methods: DNeasy Blood & Tissue Kit (manual control), Qiasymphony SP, Maxwell RSC, and KingFisher Apex (Table 1). EDTA blood extracted with QIAsymphony SP was used as an additional control to evaluate DNA integrity. DNA concentration, A260/A280 and A260/A230 ratios, DIN values, and the main-band molecular weights are shown in Table 1. DNA yields from the four extraction methods are compared in Figure 4A (Kruskal-Wallis, p=0.01). The gDNA sample with the highest yield from each method was selected for gel electrophoresis (Figure 4B). Among the automated magnetic bead-based extraction platforms, QIAsymphony SP produced the highest-quality gDNA: DNA concentration was comparable to the manual kit control, the gel band was the brightest, DIN values exceeded those of EDTA control, and TapeStation analysis showed a main peak over 60 kb (Figure S1). PCR amplification was successful (Figure 4C), confirming suitability for downstream applications. Maxwell RSC yielded gDNA with the highest concentration, slightly above manual kit control, but gel band was less intense and showed smearing (Figure 4B); DIN values and main-band molecular weight were much lower than those of the EDTA control (Figure S1), and PCR bands were relatively faint (Figure 4C). KingFisher Apex produced the lowest DNA yield, insufficient for DIN measurement; however, the main peak was>60 kb (Figure S1), and PCR amplification was successful (Figure 4C). Overall, QIAsymphony SP provides the best combination of yield, integrity, and suitability for downstream applications, while Maxwell RSC produced acceptable results and KingFisher Apex performed poorly.
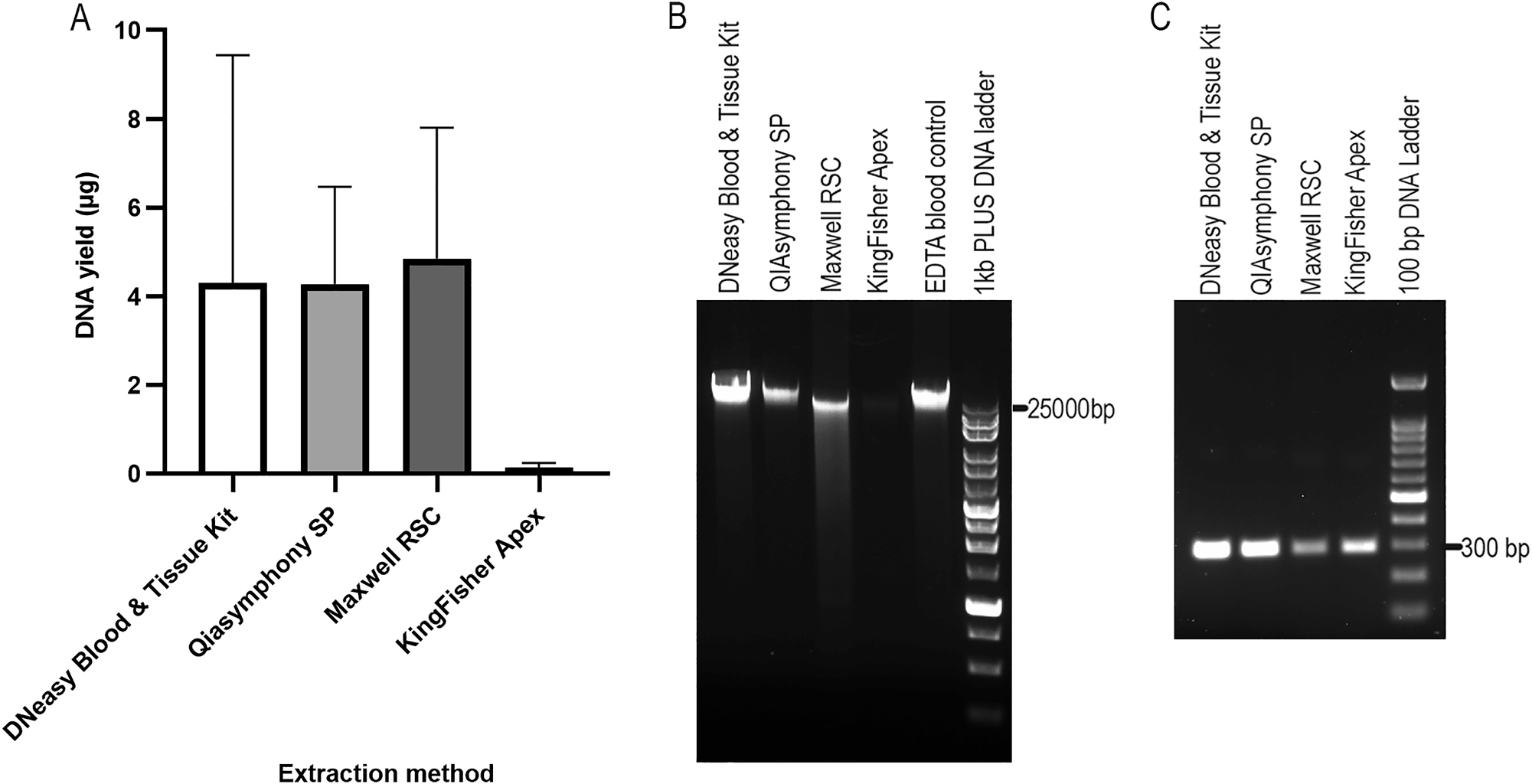
DNA yield and electrophoresis analysis of gDNA from four different extraction methods, as well as their PCR products. DNA yields are shown in (A) (n≥3, unpaired samples due to limited material, kruskal-Wallis, p=0.01). Agarose gel electrophoresis of these gDNA samples are shown in (B), with gDNA extracted from EDTA blood used as control. The PCR products (295 bp) of these gDNA samples are shown in (C).
The effect of other factors on gDNA extraction
The effect of freeze-thaw cycles on gDNA yield from PAXgene RNA blood samples
PAXgene RNA blood samples can undergo up to two freeze-thaw cycles without affecting RNA yield or stability, according to the product introductions [16]. To investigate the effect of freeze-thaw cycles on gDNA extraction, PAXgene RNA blood samples were applied to 1–5 freeze-thaw cycles (n=3). The samples were then thawed and kept at room temperature for 2 h. Pellets were collected from 1.5 mL of blood samples by centrifugation at 17000×g for 10 min. The pellet was resuspended in 450 µL of PBS, and the suspension was extracted using the QIAsymphony SP platform. As shown in Figure 5A, the highest DNA yield (3.55 ± 1.74 µg) was obtained from samples with 1 freeze-thaw cycle. Friedman test showed a significant difference between the three groups (p=0.03). Pairwise comparisons showed a significant difference between 1 freeze-thaw cycle and 5 freeze-thaw cycles (p=0.04).
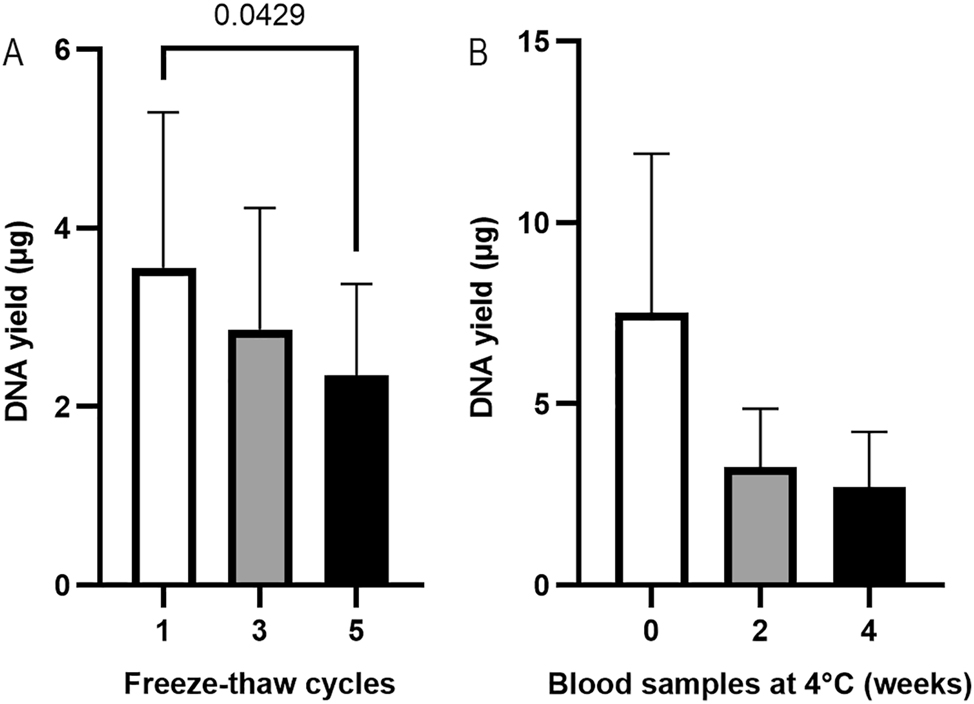
The effect of freeze-thaw cycles (A) (n=3, paired samples, Friedman test, p=0.03) and the storage at 4 °C (B) (n=3, paired samples, Friedman test, p=0.19) of the PAXgene RNA blood samples on DNA yield. gDNA was extracted using the QIAsymphony SP automated platform, and DNA yield was assessed using a NanoDrop spectrophotometer.
The effect of storage time at 4 °C on gDNA yield from PAXgene RNA blood samples
Blood samples in PAXgene blood RNA Tubes can be stored at 4 °C for up to 5 days for RNA extraction, according to the product introductions (Qiagen), whereas EDTA blood can be stored at 4 °C for months for DNA extraction [5]. To investigate the effect of storage time at 4 °C on gDNA extraction from blood samples in PAXgene Blood RNA Tubes, 1.5 mL of blood aliquots (n=3) were stored at 4 °C for 0, 2 or 4 weeks (Figure 5B). Pellets were collected from 1.5 mL of blood samples by centrifugation at 17000×g for 10 min. The pellet was resuspended in 450 µL of PBS, and the suspension was extracted using the QIAsymphony SP platform. The DNA yield from PAXgene RNA blood samples stored at 4 °C in 0, 2, 4 weeks was 7.52 ± 4.39, 3.26 ± 1.61 and 2.70 ± 1.53 µg. The highest DNA yield was obtained from blood samples stored for 0 weeks, which was about 2 folds of that of the blood samples stored in 4 °C for 2 weeks, suggesting that storage of the PAXgene RNA blood samples at 4 °C for 2 weeks should be prohibited.
Extraction of gDNA from PAXgene RNA blood samples in 96-well format
To collect additional data from clinical samples and evaluate extraction efficiency in a high-throughput format, PAXgene RNA blood samples were thawed and incubated at room temperature for 2 h. Pellets were collected from 1.5 mL of blood samples by centrifugation at 17000×g for 10 min. The pellet was resuspended in 450 µL of PBS, and the suspension was extracted using the QIAsymphony SP platform in 96-well format. The DNA yield from samples extracted using QIAsymphony SP (n=96) ranged from 0.24 to 13.46 µg, with an average of 3.92 ± 2.99 µg. A total of 12.50 % of samples had DNA yields less than 1 µg, while 66.67 % of them were within the range of 1–6 µg (Figure 6). The median value was 2.77 µg. The average A260/280 was 1.89 ± 0.89. Detailed data for each sample can be found in the Supplementary Material (Table S1). All these samples were conducted methylation assay (data not shown), which just required 500 ng DNA [17].
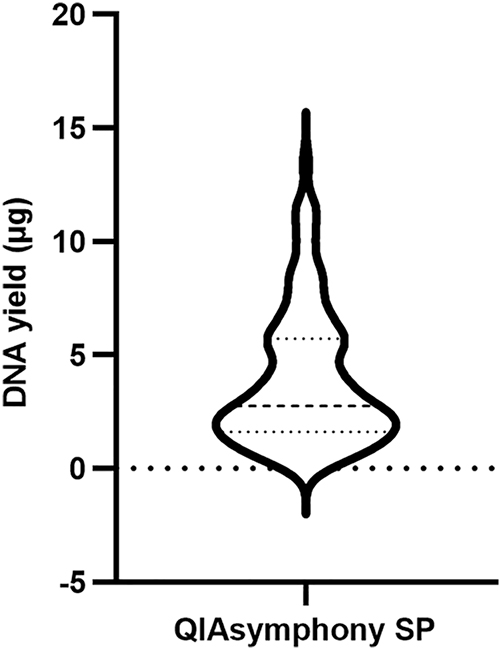
The DNA yield of PAXgene RNA blood samples extracted by QIAsymphony SP in 96-well format (n=96). The median was 2.77 µg, with the 25th and 75th percentile at 1.62 and 5.73 µg, respectively.
Discussion
EDTA blood or buffy coat is one of the best sources for gDNA extraction [18]. Although the gDNA yield from PAXgene RNA blood samples (3.92 ± 2.99 µg) in our high-throughput extractions was much lower than the gDNA yield from 400 µL of EDTA blood (30.26 ± 23.93 µg) in our previous study [5], 87.50 % of them still had a concentration above 10 ng/μL in 100 µL, which is adequate for most gDNA sequencing techniques. To our knowledge, observation of the pellet from the PAXgene RNA blood samples has not been reported. According to the product information of PAXgene Blood RNA Tube, the pellet should be lysed cells. However, it was unexpected that, although the cells were shrunken, we still could see intact cells in the pellet (Figure 1B). Many of the cells contained nuclei, which are the source of gDNA, while no nuclei were observed in the supernatant (data not shown).
Previous studies [9], [10], [11] have reported gDNA extraction from PAXgene RNA blood samples; however the specific pre-treatment details and key factors of the blood samples remain unclear or inconsistent, emphasizing the need for systematic optimization. Kelly et al. [10] centrifuged the blood sample at 4 °C, which is not recommended. According to the protocol for RNA extraction from blood samples collected in PAXgene Blood RNA Tube [15], the blood sample should be incubated at room temperature for at least 2 h and then centrifuged at room temperature before RNA extraction. Although the effect of centrifugation temperature on gDNA extraction was not evaluated in our study, it is reasonable to conclude that centrifugation at 4 °C is unnecessarily. Augello et al. [9] and Kelly et al. [10] washed cell pellet with water before DNA extraction; however, according to our results (Figure 1A), this step is unnecessary. In this study, we also investigated the effects of incubation time of blood samples at room temperature or centrifuge conditions. We found that incubating blood samples at room temperature for longer improved DNA yield (Figure 3C), and centrifugation at higher speed for longer time enhanced DNA yield as well (Figure 3A and B). However, there was no significant difference. Usually, it takes at least 1 h at room temperature to fully thaw the whole tube of PAXgene RNA blood samples (approximately 10 mL). After thawing, the whole blood can either be extracted immediately or incubated at room temperature for an additional 2 h to obtain more gDNA. If the whole blood is centrifuged at a lower speed, the pellet will be easier to resuspend. Based on our experimental results, researchers may appropriately adjust pre-treatment conditions to balance processing time and DNA yield, particularly in high-throughput applications using 96-well format.
Currently, only a few studies [9], [10], [11] have extracted gDNA from PAXgene RNA blood samples. All used silica membrane column-based methods, and obtained 2.25 µg [9], 3.75 µg [10] and 2.07 µg [11] of gDNA from 1.5 mL of PAXgene RNA blood samples (normalized), which are lower than the yields obtained with the QIAsymphony SP (4.27 ± 2.19 µg) and Maxwell RSC (4.85 ± 2.96 µg) platform in our study. This difference highlights the potential benefits of magnetic bead-based automated platforms for improving DNA recovery from PAXgene RNA blood samples. Our results indicate that the QIAsymphony SP platform consistently produced the highest-quality gDNA. Although Maxwell RSC yielded high DNA amounts, partial degradation and weaker PCR bands were observed, whereas KingFisher Apex produced significantly lower DNA yields. Notably, these findings suggest that not all magnetic bead-based kits are equally effective for extracting sufficient gDNA from PAXgene RNA blood samples. To date, only the Qiagen manual kit has been reported for gDNA extraction from PAXgene RNA blood samples. This variability suggests the importance of platform-specific validation when using any new DNA extraction kit.
In conclusion, not all magnetic bead-based platforms are suitable for gDNA extraction from PAXgene RNA blood samples. Systematic pre-treatment optimization provides important guidance for routine and high-throughput extraction workflows, and our observations on storage conditions and freeze-thaw cycles offer practical reference for biobanking.
Funding source: National Cancer Institute
Award Identifier / Grant number: P30CA016087
Acknowledgments
We would like to thank Britney Paredes Lopez, Kevin Fitzpatrick, Jonathan Kim, Cameron Stewart O’brien, Kimberly Frias, Julia Milligan and Whitey Martinez for their assistance in collecting the samples.
-
Research ethics: This study was approved by the Institutional Review Board of New York University Langone Health (IRB: S16-00122).
-
Informed consent: Informed consent was obtained from all individuals included in this study.
-
Author contributions: Jianlan You: Supervision, Investigation, Writing – original draft. Tomoe Shiomi: Investigation. Paul Zappile: Investigation. Luis Chiriboga: Writing – review & editing. Sandra Mendoza: Review & editing. Andre L. Moreira: Writing – review & editing. All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Use of Large Language Models, AI and Machine Learning Tools: ChatGPT was used only to improve English language, including grammar, spelling, and clarity, during manuscript drafting. All scientific content, data analysis, and conclusions were performed entirely by the authors. The authors reviewed and edited the content as needed after using this tool and take full responsibility for the content of the publication.
-
Conflict of interest: The authors state no conflict of interest.
-
Research funding: The NYULH Center for Biospecimen Research and Development, Molecular Laboratory (RRID: SCR_017930) and Histology and Immunohistochemistry Laboratory (RRID: SCR_018304), is supported in part by the Laura and Isaac Perlmutter Cancer Center Support Grant [NIH/ NCI P30CA016087].
-
Data availability: Data will be made available on request.
References
1. Caskey, T. Precision medicine: functional advancements. Annu Rev Med 2018;69:1–18. https://doi.org/10.1146/annurev-med-041316-090905.Search in Google Scholar PubMed
2. Murray, JR, Rajeevan, MS. Evaluation of DNA extraction from granulocytes discarded in the separation medium after isolation of peripheral blood mononuclear cells and plasma from whole blood. BMC Res Notes 2013;6:440. https://doi.org/10.1186/1756-0500-6-440.Search in Google Scholar PubMed PubMed Central
3. Gail, MH, Sheehy, T, Cosentino, M, Pee, D, Diaz-Mayoral, NA, Garcia-Closas, M, et al.. Maximizing DNA yield for epidemiologic studies: no more buffy coats? Am J Epidemiol 2013;178:1170–6. https://doi.org/10.1093/aje/kwt079.Search in Google Scholar PubMed PubMed Central
4. Lee, K, Tripathi, A. Parallel DNA Extraction from whole blood for rapid sample generation in genetic epidemiological studies. Front Genet 2020;11:374. https://doi.org/10.1039/d0sc02592e.Search in Google Scholar PubMed PubMed Central
5. You, J, Osea, J, Mendoza, S, Shiomi, T, Gallego, E, Pham, B, et al.. Automated and robust extraction of genomic DNA from various leftover blood samples. Anal Biochem 2023;678:115271. https://doi.org/10.1016/j.ab.2023.115271.Search in Google Scholar PubMed
6. Waters, J, Dhere, V, Benjamin, A, Sekar, A, Kumar, A, Prahalad, S, et al.. A practical and novel method to extract genomic DNA from blood collection kits for plasma protein preservation. J Vis Exp 2013:e4241. https://doi.org/10.3791/4241.Search in Google Scholar PubMed PubMed Central
7. Wollison, BM, Thai, E, McKinney, A, Ward, A, Clapp, A, Clinton, C, et al.. Blood collection in cell-stabilizing tubes does not impact germline DNA quality for pediatric patients. PLoS One 2017;12:e0188835. https://doi.org/10.1371/journal.pone.0188835.Search in Google Scholar PubMed PubMed Central
8. Se Fum Wong, S, Kuei, JJ, Prasad, N, Agonafer, E, Mendoza, GA, Pemberton, TJ, et al.. A simple method for DNA isolation from clotted blood extricated rapidly from serum separator tubes. Clin Chem 2007;53:522–4. https://doi.org/10.1373/clinchem.2006.078212.Search in Google Scholar PubMed
9. Augello Frank, AMRL, Matthew, WALENCIAK, Oelmueller, UWE, Wyrich, RALF, Helge, BASTIAN. Method and device for collecting and stabilizing a biological sample. Patent No.: US 6 2003;617:170. B2.Search in Google Scholar
10. Kelly, VR, Jones, SP, Sammartino, HL, Arocena, DI, Madore, SJ. Donor verification using short tandem repeat (STR) analysis directly from blood collected in PAXgene RNA tubes. Biopreserv Biobanking 2014;12:217–9. https://doi.org/10.1089/bio.2013.0080.Search in Google Scholar PubMed
11. Kruhoffer, M, Dyrskjot, L, Voss, T, Lindberg, RL, Wyrich, R, Thykjaer, T, et al.. Isolation of microarray-grade total RNA, microRNA, and DNA from a single PAXgene blood RNA tube. J Mol Diagn 2007;9:452–8. https://doi.org/10.2353/jmoldx.2007.060175.Search in Google Scholar PubMed PubMed Central
12. Fan, J, Khanin, R, Sakamoto, H, Zhong, Y, Michael, C, Pena, D, et al.. Quantification of nucleic acid quality in postmortem tissues from a cancer research autopsy program. Oncotarget 2016;7:66906–21. https://doi.org/10.18632/oncotarget.11836.Search in Google Scholar PubMed PubMed Central
13. Adewoyin, AS, Nwogoh, B. Peripheral blood film - a review. Ann Ib Postgrad Med 2014;12:71–9.Search in Google Scholar
14. Rutland, CS. Anat, J, editor. Histological and histochemical methods, 4th ed. London: Journal compilation © 2008 Anatomical Society of Great Britain and Ireland; 2008:356 p.10.1111/j.1469-7580.2008.00957.xSearch in Google Scholar
15. QIAGEN. PAXgene® Blood RNA Kit Instructions for Use (Handbook). https://www.preanalytix.com/storage/download/_ProductResources_/Handbooks/HB-0181-006_1130772_HB_PAX_Blood_RNA_0223_NA.pdf [Accesed 2023].Search in Google Scholar
16. Can the PAXgene Blood RNA Tubes undergo freeze/thaw cycles? https://www.qiagen.com/us/resources/faq/2471 [Accesed 2023].Search in Google Scholar
17. Sol, CM, Gaylord, A, Santos, S, Jaddoe, VWV, Felix, JF, Trasande, L. Fetal exposure to phthalates and bisphenols and DNA methylation at birth: the generation R study. Clin Epigenet 2022;14:125. https://doi.org/10.1186/s13148-022-01345-0.Search in Google Scholar PubMed PubMed Central
18. Stanzick, KJ, Simon, J, Zimmermann, ME, Schachtner, M, Peterhoff, D, Niller, HH, et al.. DNA extraction from clotted blood in genotyping quality. Biotechniques 2023;74:23–9. https://doi.org/10.2144/btn-2022-0061.Search in Google Scholar PubMed PubMed Central
Supplementary Material
This article contains supplementary material (https://doi.org/10.1515/cclm-2025-1079).
© 2025 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.

