Abstract
Objectives
Laboratories need to take into consideration the specificity and imprecision of assays not only in verification, but also of quality assessment. This study investigates the composition of serum used in EQA materials by comparing material from a single and multiple donors (pooled material), across multiple methods, using creatinine as an example.
Methods
Sixteen different serum matrices were distributed as 36 specimens through the UK NEQAS for Acute and Chronic Kidney Disease Scheme from March 2022 to March 2023. Male-only and female-only serum was used as single donations, pooled donations, unmanipulated or with added exogenous creatinine. Specimens were distributed to primarily UK participants (approximately n=500) for creatinine analysis. Data has been reviewed by method compared to the enzymatic creatinine method principle mean.
Results
From the 16 different matrices, only the enzymatic creatinine assay systems from Roche Cobas and Siemens Atellica met the minimum acceptable bias goal, from biological data, of 5.6 %, in all specimens. Pooled material showed less variation in bias across all methods.
Conclusions
Since Laboratories invest a lot of time and money in quality management, they need to know the limitations of their assays so that they are not investigating ‘apparent’ EQA/IQC problems which are purely due to non-specific, imprecise assay, rather than an analytical issue in their laboratory. When large numbers of individual donations are combined, interferents are essentially diluted out. Therefore, if EQA material is of this type it will be very difficult to determine the actual assay’s bias and variability.
Introduction
Modern laboratories produce a large number of test results, from a wide range of assays, that are used every day in decision making in healthcare. Therefore, in an economically and operationally constrained healthcare environment, it is ever more important that resources are used appropriately to deliver the best possible outcomes for patients. This is not only in the selection of assays before implementation, day-to-day test requesting and operational protocols, but also maintenance of the quality of service that is being provided over time. Laboratories are increasingly restricted on their choice of assay, for any analyte, as they are more often than not tied to long contracts with a single provider.
As part of ISO 15189 accreditation, laboratories are required to verify all assays before implementation [1]. This includes all parts of the manufacturers’ validation claims. Manufacturers are usually clear about their assay performance in their ‘kit inserts’ (Instructions for Use [IFU]). However, just because it is documented and the assay is CE/UKCA marked and used by other laboratories, this does not mean that it is analytically fit for the purpose that the laboratory intends to use it for. Ongoing verification of an assay’s fitness for purpose is usually maintained by regular Internal Quality Control (IQC) and External Quality Assessment (EQA). There is no single standard to which a laboratory has to perform as minimum requirements for IQC or EQA, neither is there a single standard for production of IQC or EQA material. ISO/IEC 17043:2010 (and now ISO/IEC 17043:2023) sets out standards for proficiency testing, but this does not provide definitive guidance for EQA scheme design and as such EQA schemes operate with varying number of specimens, frequency, specimen matrix and composition. This will all impact on the usefulness of the data, whether it is a snapshot of the laboratory’s performance at that point in time, or whether it reflects the overall performance as part of post market surveillance. An opinion paper was published in 2017 on the analytical performance specifications for EQA, but once again only guidance was provided [2].
EQA material can be prepared using large volumes of serum that have been pooled and manipulated to achieve different concentrations of all analytes in question covered by the EQA scheme, or alternatively single donations of serum can be dispatched with little or no manipulation. Either method of preparation can and will impact the quality of the data collected by the EQA provider which may in turn impact its usefulness and effectiveness for the laboratory.
IQC and EQA data are good sources of information about the performance of assays. Both have limitations which the laboratory needs to understand and appreciate. IQC is aimed at real time decisions, for example whether a batch of results can be released but also looks at imprecision and repeatability over a long period of time. As such, by necessity, IQC material is prepared in very large volumes to allow this consistency and so consequently running in new lots is time consuming and expensive. Just because an IQC is of a serum matrix this does not necessarily mean that it is directly comparable to clinical specimens due to the pooling of large volumes and any other manipulation that it may have undergone to ensure its stability.
There is a drive to ensure that EQA material is commutable with patient specimens, i.e. the EQA material ‘behaves’ the same way as a patient specimen would on an assay system. Formal commutability studies require the pairwise analysis of both patient specimens and EQA material on two or more methods to show that if there are the same method related biases on all materials there is commutability of the EQA material for these methods [3], [4], [5]. However, if a method is not specific or sensitive for the analyte in question, testing for the ‘commutability’ of the material in question is irrelevant as the method does not have sufficient selectivity.
Assays for creatinine have been part of the foundation of laboratory medicine for the last century [6, 7]. However, the clinical utility of the creatinine result has evolved from a simple marker of renal disease severity [8, 9] to being included in derived equations, such as estimated glomerular filtration rate (eGFR) [10, 11], and used in algorithms, such as that for acute kidney injury (AKI) where ratios are calculated with the patient’s lowest creatinine result being used as the denominator [12, 13]. Clinical trials routinely require the measurement of a subject’s renal function throughout the trial. All these situations require creatinine assays to be accurate at both high and low concentrations, have good imprecision and, crucially, be analytically stable over time.
Most manufacturers offer both a compensated kinetic Jaffe and an enzymatic creatinine assay. The limitations of the kinetic Jaffe creatinine assay, whether compensated or not, are well known and mainly relate to the poor specificity of the assay for creatinine. This means that the assay is often subject to interference not only from haemolysis, icterus and lipaemia, but also from other ‘unknown chromogens’ with similar wavelengths of absorbance and from assay architecture co-reactants such as glucose, ketones, protein etc. [14], [15], [16], [17]. However, whilst laboratories continue to purchase and routinely use the compensated kinetic Jaffe creatinine assays, manufacturers will continue to supply. There have been many publications which have examined the differences between the compensated kinetic Jaffe and enzymatic creatinine assays. The general consensus is that the enzymatic creatinine assays are preferable to their compensated kinetic Jaffe counterparts [15], [16], [17], [18] and UK NEQAS has been demonstrating and saying this since 2005.
In 2022, data from the UK NEQAS for Acute and Chronic Kidney Disease Scheme indicated that approximately 35 % of UK laboratories were still using a compensated kinetic Jaffe assay despite multiple recommendations that an assay more specific for creatinine should be used. This is even with recommendations for serum creatinine assays to be improved [19] and for laboratories to use creatinine assays that are specific for creatinine (for example, enzymatic assays) and zero-biased compared with isotope dilution mass spectrometry (IDMS) [20]. It should be pointed out that enzymatic creatinine assays are not without their own limitations and there have been a number of publications highlighting these, for example interference from high concentrations of lithium heparin (under-filled tubes) [21], high concentrations of N-acetylcysteine [22], IgM paraproteinaemia [23] and haemolysis [24].
Laboratories can, and do, spend a considerable amount of time and money performing IQC and EQA and they should be taking their choice of assay and any specificity issues into consideration when evaluating assay bias and imprecision. An assay can only ever be as good as what it was designed for. This is true for all assays and laboratories should be risk assessing these limitations for patients in terms of the population that they serve.
The purpose of this study was to investigate the composition of serum used in EQA materials by comparing material from a single donor, multiple donors (pooled material) and the spiking of a single analyte into pooled material, across multiple methods. Creatinine has been used as an example due to the clinical importance of this assay and the wide application of creatinine results. The UK NEQAS for Acute and Chronic Kidney Disease Scheme was utilised for the study as it allowed a large number of laboratories and methodologies to be included over a number of matrices and samples.
Materials and methods
Specimen preparation
From March 2022 (Distribution 185) to March 2023 (Distribution 196) a total of 36 liquid, serum specimens were distributed for creatinine analysis as part of the UK NEQAS for Acute and Chronic Kidney Disease EQA Scheme for the determination of eGFR.
All serum was sourced either as off-the-clot serum from NHS Blood and Transplant, Filton, UK or commercially procured from TCS Biosciences Ltd., Buckingham, UK. Serum was received frozen and either used as an ‘individual donation’ (after being filtered to 1 um, using a glass fibre filter paper, Merck, Gillingham, UK), or used as ‘pooled material’ with serum donations from donors with the same blood group being mixed together, and then filtered as above [Using serum from the same blood group is one of Birmingham Quality’s protocols to minimise the potential of micro clots which could impact a laboratory’s analytical result.] Higher concentrations of creatinine were achieved by the addition of anhydrous creatinine, Merck, Gillingham, UK (up to a maximum concentration of ∼300 μmol/L) and during this time period one specimen was diluted using 5 % w/v human serum albumin (HSA) (Pharmacy Grade).
Six individual donations (three male and three female) were distributed, un-manipulated, as six of the 36 specimens. Ten different serum ‘pools’ (four male and six female) were distributed as the remaining 30 specimens. These ‘pools’ were either un-manipulated (n=4), had added creatinine (n=25) and one specimen was diluted with 5 % w/v Human Serum Albumin. Pool volumes ranged from ∼1.25 L (eight individual donations) to ∼4.5 L (30 individual donations). In total there were 16 different serum matrices distributed throughout the year. This frequency of EQA specimens distributed is at the high end of what is typically used in Europe and the USA.
Three specimens are distributed, notionally monthly, to all participants enrolled in the EQA Scheme. Specimens are stored frozen until dispatch where they were then transported at room temperature (the majority of which arrive within 24 h). Participants are advised to analyse immediately as if they were from a patient. For each specimen, participants report back online (manually or automatically), to the Scheme at Birmingham Quality, the creatinine concentration determined. Participants are also able to check the method that they are using and can change if required to ensure that they are registered for the correct method.
Data analysis
In the UK NEQAS for Acute and Chronic Disease scheme outliers are removed by Healy trimming which is applied to all data [25]. This essentially leads to the highest and lowest 5 % of specimen level data being removed from statistical calculations of means and CVs.
Creatinine results are evaluated at both method principle (for example compensated kinetic Jaffe or enzymatic) and at the manufacturer analyser level (Abbott Alinity, Roche Cobas etc.). A mean, standard deviation and coefficient of variation is calculated for each method. However, the target value for every laboratory, irrespective of the method that they are using for creatinine, is the enzymatic creatinine method principle mean, being calculated from results from specific methods. The UK NEQAS for Acute and Chronic Kidney Disease scheme uses the enzymatic creatinine principle as the method mean for a number of reasons (1) the results are available in real time and (2) the number of specimens that are distributed (three, monthly) would make it a very expensive EQA service if every specimen had a reference method value. The enzymatic creatinine principle method means are periodically checked by the analysis of specimens using a Reference Method accredited to ISO/IEC 17025:2017 to give further confidence in the targets – and there is good agreement – therefore there is no justification for undertaking this additional work. This is continually reviewed and if the enzymatic creatinine assays were to change the Scheme would also change to reflect this.
Results
Accuracy
The target value for creatinine in the UK NEQAS for Acute and Chronic Kidney Disease Scheme is the enzymatic creatinine method principle mean and approximately 250 results have contributed to it for each of the 36 specimens. The enzymatic creatinine method principle mean has been validated, during this time period, by analysis of 16 specimens by an isotope-dilution mass spectrometry reference method. The correlation between the enzymatic creatinine method mean and the reference method has an R2 of 0.9989, and at a creatinine concentration of 100 μmol/L the difference between the two is only 0.68 % (100.7 μmol/L).
Figure 1 shows by method, the % bias for each specimen from Distribution 185 to Distribution 196. This data and a summary is also tabulated in Table 1. Table 1 shows details for each specimen as well as the method bias for each specimen compared to the enzymatic creatinine target value. The summary at the bottom of the table gives the minimum, maximum and median number of data points that have contributed to the method mean which the bias has been calculated from, as well as the minimum, maximum and median biases. The number of individual specimens and individual matrices where the minimum acceptable bias for creatinine of 5.6 % is also given. The minimum acceptable bias for creatinine using analytical performance specification data from biological variation is 5.6 %. This has been chosen as this is the performance specification that has been agreed and is used in the UK NEQAS for Acute and Chronic Kidney Disease scheme. Though analytically it may be better to use desirable analytical performance specifications from biological variation data or even outcome study data, pragmatically we have found that participant laboratories do not engage with us if their targets and goals are perceived to be unobtainable or have to be met in a single step [26].

Bias plot for serum creatinine by method for specimens distributed from March 2022 (Distribution 185) to March 2023 (Distribution 196). The minimum acceptable bias for creatinine using analytical performance specification data from biological variation is 5.6 %.
Summary table of method mean % biases for 36 specimens distributed from March 2022 (Distribution 185) to March 2023 (Distribution 196). Specimen 188C (in italics) is a pool composed of serum diluted with human serum albumin; all other specimens have 100 % serum as their base material.
| Specimen number | Pool series # | Gender | Single donation [S] | Pooled material [P] | Spike [X] | Enzymatic target value | Abbott Alinity [10AB20] | Abbott Architect [10AB] | Beckman AU [10OL] | Roche Cobas [10BO] | Siemens ADVIA [10TE] | Siemens Atellica [10SM20] | Abbott Alinity [9AB20] | Abbott Architect [9AB] | Beckman AU [9OL] | Roche Cobas [9BO] | Siemens ADVIA [9TE] | Siemens Atellica [9SM20] | Specimen number |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 185A | #1 | M | P | X | 135.8 | −2.7 | 0.0 | −4.7 | −0.5 | −3.9 | −4.1 | −4.3 | 2.2 | 1.7 | −0.7 | 1.8 | 185A | ||
| 185B | #1 | M | P | X | 159.8 | −3.1 | −1.2 | −5.5 | −1.5 | −4.4 | −4.1 | −4.1 | 1.7 | 1.8 | −0.7 | 1.7 | 185B | ||
| 185C | #1 | M | P | X | 292.8 | −1.4 | −0.5 | −5.2 | −3.6 | −6.1 | −2.8 | −3.1 | 0.6 | 1.3 | 0.0 | 0.7 | 185C | ||
| 186A | F | S | 50.1 | 8.4 | 13.4 | 1.4 | 6.6 | −2.7 | −2.6 | −3.5 | 3.6 | 1.2 | −3.2 | −1.7 | 186A | ||||
| 186B | F | S | 88.6 | −3.8 | 0.5 | 2.4 | 5.7 | −0.5 | −3.1 | −4.2 | 2.7 | 1.7 | −2.9 | −1.9 | 186B | ||||
| 186C | M | S | 93.3 | 1.6 | 5.7 | −3.6 | 6.4 | 2.2 | −4.2 | −3.9 | 2.9 | 1.8 | −3.0 | −0.3 | 186C | ||||
| 187A | #2 | M | P | X | 122.3 | −1.5 | −0.2 | −3.8 | −0.7 | −2.5 | −3.6 | −3.6 | 3.4 | 1.2 | −0.5 | 0.0 | 187A | ||
| 187B | #2 | M | P | X | 132.4 | −1.5 | −0.6 | −3.6 | −1.1 | −3.7 | −3.5 | −3.5 | 3.2 | 1.1 | −1.1 | 0.7 | 187B | ||
| 187C | #2 | M | P | X | 287.4 | −1.2 | 0.0 | −3.7 | −3.5 | −6.1 | −1.9 | −2.1 | 2.2 | 0.6 | −0.9 | 0.0 | 187C | ||
| 188A | #3 | F | P | 66.9 | 2.0 | 1.7 | −3.2 | 6.6 | 0.1 | −1.4 | −3.2 | −1.3 | 2.2 | −13.0 | −0.7 | 188A | |||
| 188B | #3 | F | P | X | 274.9 | −1.3 | 0.2 | −3.1 | −2.3 | −6.3 | −1.3 | −1.9 | 0.7 | 1.0 | −3.5 | −0.2 | 188B | ||
| 188C* | #3 | F | P | X | 32.1 | 26.9 | 33.3 | −3.8 | 8.9 | −0.6 | −0.5 | −2.3 | 6.0 | 0.1 | −8.7 | 1.3 | 188C* | ||
| 189A | #4 | F | P | X | 73.7 | −2.2 | −2.8 | −2.7 | 4.6 | −1.2 | −1.7 | −3.1 | 0.4 | 1.7 | −6.6 | −0.6 | 189A | ||
| 189B | #4 | F | P | X | 116.3 | −3.8 | −2.3 | −3.0 | 0.8 | −4.4 | −1.8 | −2.9 | 0.7 | 1.6 | −4.8 | −0.8 | 189B | ||
| 189C | #4 | F | P | X | 142.8 | −2.9 | −1.3 | −3.3 | −0.5 | −4.4 | −1.7 | −2.7 | 1.0 | 1.4 | −4.3 | −0.8 | 189C | ||
| 190A | F | P | 65.0 | −0.3 | 0.9 | −0.5 | 7.4 | −1.5 | −1.6 | −2.4 | 4.1 | 0.7 | −3.6 | −0.1 | 190A | ||||
| 190B | F | P | 66.7 | −1.8 | 0.8 | −1.4 | 7.9 | 0.7 | −2.0 | −2.4 | 2.0 | 1.3 | −4.8 | 0.1 | 190B | ||||
| 190C | M | P | 88.4 | −3.8 | −1.4 | −9.0 | 2.7 | 0.2 | −3.5 | −4.5 | 1.1 | 2.3 | −5.3 | 1.5 | 190C | ||||
| 191A | #5 | F | P | X | 89.2 | −5.6 | −4.0 | −1.2 | 3.5 | −3.3 | −2.5 | −3.3 | 3.0 | 1.1 | −2.1 | −0.6 | 191A | ||
| 191B | #5 | F | P | X | 121.2 | −4.4 | −3.0 | −2.2 | 1.1 | −5.0 | −2.1 | −3.2 | 1.7 | 1.3 | −2.9 | −0.9 | 191B | ||
| 191C | #5 | F | P | X | 173.1 | −3.4 | −1.9 | −2.9 | −0.8 | −6.1 | −2.1 | −2.5 | 1.9 | 1.1 | −2.1 | −0.7 | 191C | ||
| 192A | #2 | M | P | X | 124.1 | −1.7 | −1.7 | −5.1 | −1.5 | −2.1 | −3.9 | −4.5 | −5.1 | 3.5 | 2.0 | −1.8 | 2.3 | 192A | |
| 192B | #2 | M | P | X | 134.4 | −3.5 | −2.8 | −4.7 | −2.4 | −2.6 | −4.4 | −4.4 | −4.9 | 3.5 | 2.0 | −1.9 | 1.6 | 192B | |
| 192C | #2 | M | P | X | 289.6 | −1.2 | −0.6 | −4.8 | −4.4 | −5.4 | −6.8 | −2.9 | −2.2 | 2.5 | 1.1 | −1.4 | 0.6 | 192C | |
| 193A | #3 | F | P | X | 77.8 | −7.3 | −2.7 | −3.1 | 2.4 | −2.2 | 1.4 | −3.6 | −4.1 | −1.1 | 2.7 | −8.7 | −1.2 | 193A | |
| 193B | #3 | F | P | X | 118.8 | −5.8 | −2.6 | −5.0 | −0.4 | −5.2 | −4.1 | −3.5 | −3.1 | 0.6 | 2.1 | −5.8 | −1.0 | 193B | |
| 193C | #3 | F | P | X | 222.6 | −3.8 | −0.6 | −4.4 | −2.9 | −5.7 | −5.7 | −2.9 | −2.0 | 1.2 | 1.4 | −3.2 | −0.5 | 193C | |
| 194A | #6 | M | P | X | 114.0 | −2.3 | −0.5 | −7.1 | −0.8 | −1.7 | 0.3 | −4.4 | −3.3 | 1.7 | 1.7 | −3.1 | 1.4 | 194A | |
| 194B | #6 | M | P | X | 136.6 | −1.9 | 0.2 | −6.9 | −1.6 | −2.1 | −1.1 | −4.6 | −4.4 | 2.0 | 2.0 | −3.3 | 1.3 | 194B | |
| 194C | #6 | M | P | X | 238.9 | −0.7 | −0.5 | −5.9 | −3.6 | −4.7 | −4.6 | −2.8 | −1.9 | 1.2 | 1.2 | −2.0 | 0.2 | 194C | |
| 195A | M | S | 94.2 | 13.9 | 11.6 | −2.3 | 5.6 | 11.2 | 14.4 | −6.4 | −9.2 | 2.7 | 2.8 | −3.5 | 0.3 | 195A | |||
| 195B | F | S | 76.4 | −8.9 | −5.8 | −3.5 | −1.0 | −7.0 | −3.0 | −3.6 | −3.3 | 5.5 | 1.1 | −3.3 | −1.8 | 195B | |||
| 195C | M | S | 68.8 | −5.0 | −4.9 | −8.5 | −0.3 | −2.8 | 1.7 | −5.5 | −6.6 | 5.7 | 1.7 | −2.1 | 0.5 | 195C | |||
| 196A | #7 | F | P | X | 80.7 | −4.3 | −2.5 | −0.2 | 4.6 | −0.1 | 2.7 | −3.1 | −3.0 | −0.6 | 1.7 | −6.5 | −2.7 | 196A | |
| 196B | #7 | F | P | X | 121.2 | −5.1 | −0.3 | −0.8 | 1.2 | −2.4 | −0.2 | −2.9 | −3.3 | −0.3 | 1.6 | −5.6 | −2.1 | 196B | |
| 196C | #7 | F | P | X | 171.8 | −4.2 | −0.5 | −1.1 | −0.7 | −4.4 | −3.4 | −2.5 | −2.7 | 0.0 | 1.5 | −4.5 | −1.5 | 196C | |
|
|
|||||||||||||||||||
| Enzymatic target value | Abbott Alinity [10AB20] | Abbott Architect [10AB] | Beckman AU [10OL] | Roche Cobas [10BO] | Siemens ADVIA [10TE] | Siemens Atellica [10SM20] | Abbott Alinity [9AB20] | Abbott Architect [9AB] | Beckman AU [9OL] | Roche Cobas [9BO] | Siemens ADVIA [9TE] | Siemens Atellica [9SM20] | |||||||
|
|
|||||||||||||||||||
| Min ‘n’ | 9 | 8 | 11 | 76 | 10 | 7 | 45 | 29 | 22 | 154 | 11 | 12 | |||||||
| Max ‘n’ | 17 | 15 | 15 | 88 | 17 | 10 | 56 | 38 | 32 | 172 | 20 | 15 | |||||||
| Median ‘n’ | 13 | 14 | 13 | 84 | 15 | 10 | 49 | 33 | 30 | 159 | 17 | 15 | |||||||
| Min | 32 | −8.9 | −5.8 | −9.0 | −4.4 | −7.0 | −6.8 | −6.4 | −9.2 | −1.3 | 0.6 | −13.0 | −1.9 | ||||||
| Max | 293 | 13.9 | 13.4 | 2.4 | 7.9 | 11.2 | 14.4 | −1.3 | −1.9 | 5.7 | 2.8 | 0.0 | 2.3 | ||||||
| Median | 120 | −2.3 | −0.6 | −3.6 | −0.5 | −3.1 | −3.4 | −3.0 | −3.3 | 2.0 | 1.5 | −3.1 | −0.1 | ||||||
-
Any bias values outside the 5.6 % limits have been printed in bold italics.
The enzymatic creatinine assays have a more consistent bias across the concentration range (the bias varies between manufacturers), whereas the compensated kinetic Jaffe creatinine assays show more variation in bias at lower concentrations.
All compensated kinetic Jaffe creatinine assays had at least one specimen that exceeded the 5.6 % minimum bias with 6/16 serum matrices being impacted for both the Roche Cobas and Siemens Advia compensated kinetic Jaffe assays. The enzymatic creatinine assays showed less bias with both the Roche Cobas and Siemens Atellica enzymatic assays having method means for all 16 serum matrices within the 5.6 % minimum bias. There were some individual specimens where the bias was within 5.6 % for all method/manufacturer combinations, but this was only ever seen in pooled material, for example Specimen 185A, 187B and 196C.
Imprecision
Imprecision profiles for all methods from Distribution 185 to Distribution 196 are shown in Figure 2. The minimum acceptable analytical imprecision for creatinine using analytical performance specification data from biological variation is 3.4 % [26] and this is highlighted on each graph. The imprecision is numerically lower for the enzymatic assays across the concentration range (0–300 μmol/L). A minimum number of data points of seven were used to calculate analytical imprecision; the maximum number of data points was over 170.

Imprecision profiles for serum creatinine for all methods from March 2022 (Distribution 185) to March 2023 (Distribution 196). The minimum acceptable analytical imprecision for creatinine using analytical performance specification data from biological variation is 3.4 %.
Single donations
Six of the specimens were derived from single donations of serum (Attribute S – 186A, 186B, 186C, 195A, 195B and 195C). Though formal commutability studies have not been performed, these are considered the most representative material compared to routine clinical specimens. Reference Values were obtained on all six of these donations (creatinine concentration 51.0–93.4 μmol/L) and in all cases there was good agreement with the enzymatic creatinine method mean (R2 of 0.9972). None of the six donations showed every manufacturer method mean to be within the minimum acceptable bias of 5.6 % (Table 1). Four out of the six donations did met the minimum acceptable bias for all manufacturer’s enzymatic creatinine methods. One limitation of the study is that creatinine was not added to single donations. This was because we wanted material to be used in its most native form, but this has limited evaluation of data with regard to added creatinine.
Pooled donations
Pooled material was distributed with or without added creatinine. In total, four different pools of serum without added creatinine (Attribute P) and seven different pools with added creatinine (Attribute PX – the base for one of these pools was distributed as one of the endogenous pools) were distributed.
Review of the data from the four different endogenous creatinine only pools (188A, 190A, 190B and 190C) showed the Roche Cobas compensated kinetic Jaffe creatinine assay failed to meet the minimum acceptable bias of 5.6 % in three out of four of the specimens, the Beckman AU compensated kinetic Jaffe creatinine assay failed on one specimen and the Siemens ADVIA enzymatic creatinine assay failed on one specimen.
From the seven different pooled materials that had different amounts of creatinine added, no single material acheived the minimum acceptable bias criteria for all assays. Overall there is less of an impact on bias in the pooled material with added creatinine, but this is likely to be because the creatinine concentration is higher and assays have better imprecision at higher concentrations.
Figure 3 shows relative method bias plotted graphically for three single donations (195A [A], 195B [B] and 195C [C]), three different pooled donations (190A [D], 190B [E] and190C [F]) and three pools from the same pooled base with added creatinine (192A [G], 192B [H] and 192C [I]). It can be seen that there is a higher degree of variation between methods, both for compensated kinetic Jaffe and enzymatic method principles, in the three single donations of ‘normal creatinine concentration’ relative to pooled material also of ‘normal creatinine concentration’. When creatinine is added into serum to achieve higher concentrations (Figure 3(C)) the relative method bias is mainly related to the base material, not the added creatinine. We know that the imprecision of creatinine assays decreases at higher concentrations, therefore this may explain why there is little impact on relative method bias as creatinine concentration increases.

Relative method bias for (A–C) three individual single donations, (D–F) three different pooled donations and (G–I) three pools from the same pooled base with added creatinine. Each individual method bias has been calculated against the enzymatic creatinine method mean.
Reagent variability
Though we did not collect reagent/calibrator lot number information from Participants for their creatinine assay, it can be assumed that there have would been changes to both over the time period of the study. There may have been multiple lots of reagent in use at any one time for any/all manufacturers. Figure 4 shows for each manufacturer the box and whisker spread of B Score (Bias Score – the average of 18 individual specimen %biases over a rolling time window of six months) against time, which is represented as the distribution number on the x-axis. Notionally the distributions are one month apart, therefore the x-axis covers a five year period. Plots such as those shown in Figure 4 give an indication of the spread and variation in biases for individual methods at any one time. It is likely that it is the differences in reagent/calibrator lots that contribute to this. For some manufacturer/assay combinations, such as the Roche Cobas enzymatic creatinine assay, the assay bias and spread or participant results has remained relatively stable; whereas the Siemens ADVIA enzymatic creatinine assay has shown a change in B score, from >−9 % to approximately −3 % at Distribution 196 (March 2023). The spread of data for the Siemens ADVIA compensated kinetic Jaffe creatinine assay is much tighter at Distributions 192–196 than those previous.
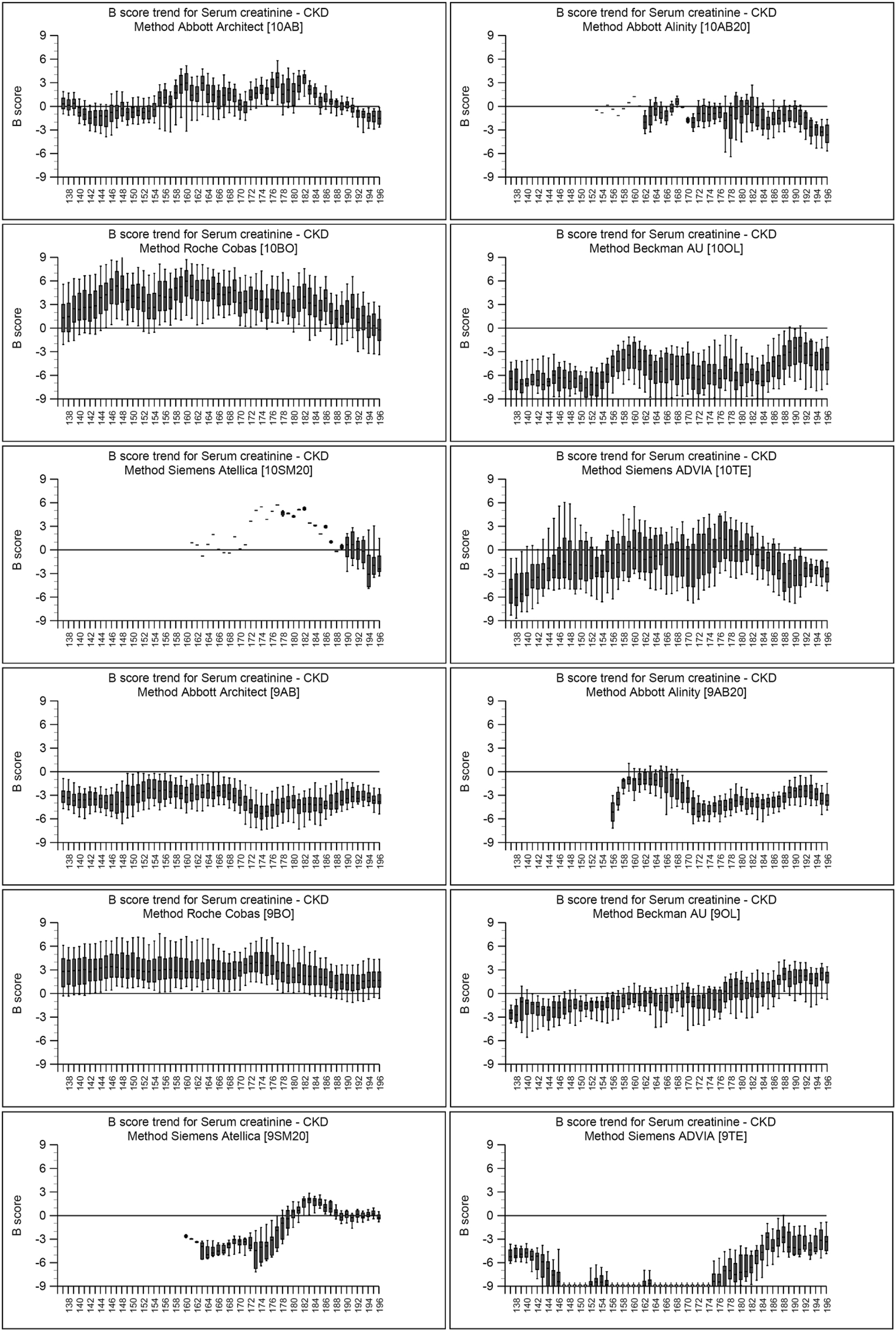
Box and Whisker B score trend plots for serum creatinine for all methods from October 2017 (Distribution 137) to March 2023 (Distribution 196). The B score is the bias score calculated from the average of 18 individual specimen biases over a six month time period.
Discussion
Through the UK NEQAS for Acute and Chronic Kidney Disease Scheme, over the 12 month period March 2022 to March 2023, 36 specimens have been distributed. These were as 16 different serum matrices either as single donations, pooled donations or pooled donations with added creatinine. Variation in relative method biases has been observed which is not consistent between methods on different serum base material. This is evident on both single and pooled donations; however, there is some consistency when the same base material is used with different amounts of creatinine added.
As expected, the compensated kinetic Jaffe assays are those mainly impacted and though this is not a new phenomenon, it is disappointing that laboratories are continuing to purchase and use these assays. Those laboratories need to be aware that the specificity deficiencies of those assays will impact IQC, EQA as well as their patients. Laboratories should always be taking into consideration their service requirements and focus on the clinically relevant concentrations, for them. In the case of creatinine, good accuracy and reproducibility at low concentrations is just as important, or potentially more important, than that at elevated concentrations as these results are included as a denominator in algorithmic calculations such as that for AKI.
It is impossible to routinely identify inaccurate results in patient specimens; however, IQC and falsely ascribed EQA ‘errors’ will be identified and could lead to unnecessary investigation which is both costly financially and in laboratory time. The laboratory needs to be aware of the limitations of all assays that they are using and take these into account in EQA review, like they would for their risk assessment in patient management.
In this study, lack of commutability of EQA material cannot be excluded as no formal commutability studies have been undertaken; however, minimally manipulated fresh frozen human serum is more likely to behave as clinical specimens compared to say delipidated, defibrinated plasma. The use of frozen material could still be considered a limitation of the study as patient specimens are likely to be analysed fresh.
Use of pooled serum which has been highly manipulated may artificially give false reassurance about the performance of the assay which would not be replicated in patient specimens due to the dilution of any unnknown chromogens with similar wavelengths of absorbance or other interfering compounds. The laboratory needs to be aware of assay specificity and imprecision issues and should consider the type of base material(s) that are provided in EQA schemes as well as the frequency and concentration range covered. Interfering substances have not been discussed in depth as part of this work, but the laboratory should appreciate they are not theoretical possibilities, they are a reality in the clinical specimens in laboratories. A single base material which has been highly manipulated or a low frequency scheme, will not adequately assess the quality and performance of their assays. This is not only crucial for an individual laboratory, but also forms the basis of post market surveillance which is essential for all laboratories to ensure that only the best and most suitable assays are available on the market.
Though this paper uses creatinine as an example the generic principles that have been discussed with regard to EQA Scheme design hold true for all analytes and methods. ISO 15189:2022 is written as a risk based approach for obtaining the best for patient care. EQA as one of the tools of quality management should be fully utilised, where appropriate with well designed schemes, to assist with risk management and not seen as an additional parallel process.
Conclusions
It is only the intensive scheme design that allows these types of observation to be made. It would be impossible for a low frequency scheme (because of insufficient data points) or a large participant scheme (where sample volumes mean that single donation or oligo donation pools cannot be used) to tease out these subtleties. Similarly, any scheme that has a large number of analytes for evaluation within a single specimen would struggle to be definitive for all analytes (the matrix would be a compromise). Although having the luxury of having a single measurand per tube, as some EQA Schemes do for Steroid Hormones, it is not a practical proposition for most situations, nevertheless having everything in the same tube is convenient but ultimately not satisfying as there are too many confounding factors to make the data definitive. EQA Organisers need to be cognisant of the limitations of their matrices and Scheme design, and many are. However, laboratories have a responsibility to actively choose to participate in EQA Schemes that give a genuine insight into assay performance. Analysis of unchallenging, highly processed samples is false economy as this does not provide any additional information to the quality of assays and can lead to resource being wasted investigating so called ‘EQA errors’.
-
Research ethics: Not applicable.
-
Informed consent: Not applicable.
-
Author contributions: The authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Competing interests: The authors state no conflict of interest.
-
Research funding: None declared.
-
Data availability: Not applicable.
References
1. International Organisation for Standardization. ISO 15189 medical laboratories – requirements for quality and competence. Geneva: ISO; 2012.Search in Google Scholar
2. Jones, GRD, Albarede, S, Kesseler, D, MacKenzie, F, Mammen, J, Pedersen, M, et al.. Analytical performance specifications for external quality assessment – definitions and descriptions. Clin Chem Lab Med 2017;55:949–55. https://doi.org/10.1515/cclm-2017-0151.Search in Google Scholar PubMed
3. Braga, F, Panteghini, M. Commutability of reference and control materials: an essential factor for assuring the quality of measurements in laboratory medicine. Clin Chem Lab Med 2019;57:967–73. https://doi.org/10.1515/cclm-2019-0154.Search in Google Scholar PubMed
4. Miller, WG, Schimmel, H, Rej, R, Greenberg, N, Ceriotti, F, Burns, C, et al.. IFCC Working Group recommendations for assessing commutability part 1: general experimental design. Clin Chem 2018;64:447–54. https://doi.org/10.1373/clinchem.2017.277525.Search in Google Scholar PubMed PubMed Central
5. Sandberg, S, Fauskanger, P, Johansen, JV, Keller, T, Budd, J, Greenberg, N, et al.. Recommendations for setting a criterion and assessing commutability of sample materials used in external quality assessment/proficiency testing schemes. Clin Chem 2023;69:1227–37. https://doi.org/10.1093/clinchem/hvad135.Search in Google Scholar PubMed
6. Delanghe, JR, Speeckaert, MM. Creatinine determination according to Jaffe-what does it stand for? NDT Plus 2011;4:83–6. https://doi.org/10.1093/ndtplus/sfq211.Search in Google Scholar PubMed PubMed Central
7. Roscoe, MH. The estimation of creatinine in serum. J Clin Pathol 1953;6:201–7. https://doi.org/10.1136/jcp.6.3.201.Search in Google Scholar PubMed PubMed Central
8. Goldman, R. The clinical evaluation of renal function. Calif Med 1956;85:376–80.Search in Google Scholar
9. Enger, E, Blegen, EM. The relationship between endogenous creatinine clearance and serum creatinine in renal failure. Scand J Clin Lab Invest 1964;16:273–80. https://doi.org/10.1080/00365516409060517.Search in Google Scholar PubMed
10. Levey, AS, Bosch, JP, Lewis, JB, Greene, T, Rogers, N, Roth, D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med 1999;130:461–70. https://doi.org/10.7326/0003-4819-130-6-199903160-00002.Search in Google Scholar PubMed
11. Levey, AS, Stevens, LA, Schmid, CH, Zhang, YL, Castro, AF3rd, Feldman, HI, et al.. A new equation to estimate glomerular filtration rate. Ann Intern Med 2009;150:604–12. https://doi.org/10.7326/0003-4819-150-9-200905050-00006.Search in Google Scholar PubMed PubMed Central
12. Algorithm for detecting Acute Repeat Kidney Injury (AKI) based on serum creatinine changes with time. UK: NHS England; 2014. https://www.england.nhs.uk/wp-content/uploads/2014/06/psa-aki-alg.pdf [Accessed 23 Jan 2022].Search in Google Scholar
13. Marrington, R, Barton, AL, Yates, A, McKane, W, Selby, NM, Murray, JS, et al.. National recommendations to standardise acute kidney injury detection and alerting. Ann Clin Biochem 2023;60:406–16. https://doi.org/10.1177/00045632231180403.Search in Google Scholar PubMed
14. Peake, M, Whiting, M. Measurement of serum creatinine--current status and future goals. Clin Biochem Rev 2006;27:173–84.Search in Google Scholar
15. Drion, I, Cobbaert, C, Groenier, KH, Weykamp, C, Bilo, HJ, Wetzels, JF, et al.. Clinical evaluation of analytical variations in serum creatinine measurements: why laboratories should abandon Jaffe techniques. BMC Nephrol 2012;13:133. https://doi.org/10.1186/1471-2369-13-133.Search in Google Scholar PubMed PubMed Central
16. Greenberg, N, Roberts, WL, Bachmann, LM, Wright, EC, Dalton, RN, Zakowski, JJ, et al.. Specificity characteristics of seven commercial creatinine measurement procedures by enzymatic and Jaffe method principles. Clin Chem 2012;58:391–401. https://doi.org/10.1373/clinchem.2011.172288.Search in Google Scholar PubMed
17. Delanaye, P, Cavalier, E, Pottel, H. Serum creatinine: not so simple! Nephron 2017;136:302–8. https://doi.org/10.1159/000469669.Search in Google Scholar PubMed
18. Hoste, L, Deiteren, K, Pottel, H, Callewaert, N, Martens, F. Routine serum creatinine measurements: how well do we perform? BMC Nephrol 2015;16:21. https://doi.org/10.1186/s12882-015-0012-x.Search in Google Scholar PubMed PubMed Central
19. Myers, GL, Miller, WG, Coresh, J, Fleming, J, Greenberg, N, Greene, T, et al.. Recommendations for improving serum creatinine measurement: a report from the Laboratory Working Group of the National Kidney Disease Education Program. Clin Chem 2006;52:5–18. https://doi.org/10.1373/clinchem.2005.052514.Search in Google Scholar
20. National Institute for Health and Care Excellence. Chronic kidney disease in adults: assessment and management guidelines. NICE Guideline [CG182]. UK: NICE; 2014.Search in Google Scholar
21. Squires, M, Wise, H, Holmes, H, Hadfield, K. Lithium heparin interference in the Abbott enzymatic creatinine assay: the significance of under-filled tubes. Ann Clin Biochem 2021;58:653–6. https://doi.org/10.1177/00045632211040673.Search in Google Scholar PubMed
22. Huang, JW, Lahey, B, Clarkin, OJ, Kong, J, Clark, E, Kanji, S, et al.. A systematic review of the effect of N-acetylcysteine on serum creatinine and cystatin C measurements. Kidney Int Rep 2021;6:396–403. https://doi.org/10.1016/j.ekir.2020.11.018.Search in Google Scholar PubMed PubMed Central
23. Flowers, KC, Tuddenham, E, Leiva, A, Garrison, L, Morris, JE, Cromwell, T, et al.. Negative interference from immunoglobulin M paraproteinaemia on the Roche enzymatic creatinine method. Ann Clin Biochem 2022;59:205–10. https://doi.org/10.1177/00045632221074867.Search in Google Scholar PubMed
24. Birmingham Quality (UK NEQAS). UK NEQAS for serum indices distribution 142. Birmingham: Birmingham Quality; 2020.Search in Google Scholar
25. Healy, MJ. Outliers in clinical chemistry quality-control schemes. Clin Chem 1979;25:675–7. https://doi.org/10.1093/clinchem/25.5.675.Search in Google Scholar
26. Aarsand, AKF-CP, Webster, C, Coskun, A, Gonzales-Lao, E, Diaz-Garzon, J, Jonker, N, et al.. The EFLM biological variation database. https://biologicalvariation.eu/ [Accessed 15 Apr 2023].Search in Google Scholar
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorials
- EFLM European Urinalysis Guideline
- Clinical Chemistry Laboratory Medicine in the post-acute COVID-19 era
- EFLM Guideline
- The EFLM European Urinalysis Guideline 2023
- Review
- Approaching sustainability in Laboratory Medicine
- Opinion Papers
- New reimbursement models to promote better patient outcomes and overall value in laboratory medicine and healthcare
- Screening for sickle cell disease: focus on newborn investigations
- Genetics and Molecular Diagnostics
- The role of Killer immunoglobulin-like receptors (KIRs) in the genetic susceptibility to non-celiac wheat sensitivity (NCWS)
- General Clinical Chemistry and Laboratory Medicine
- Understanding the limitations of your assay using EQA data with serum creatinine as an example
- Add-on testing: stability assessment of 63 biochemical analytes in centrifuged and capped samples stored at 16 °C
- Smartphone swabs as an emerging tool for toxicology testing: a proof-of-concept study in a nightclub
- Allergy: Evaluation of 16 years (2007–2022) results of the shared external quality assessment program in Belgium, Finland, Portugal and The Netherlands
- Cancer Diagnostics
- Monoclonal whole IgG impairs both fibrin and thrombin formation: hemostasis and surface plasmon resonance studies
- Cardiovascular Diseases
- Patients with antiphospholipid syndrome and a first venous or arterial thrombotic event: clinical characteristics, antibody profiles and estimate of the risk of recurrence
- Letters to the Editor
- Familial dysalbuminemic hyperthyroxinemia coexisting with a Grave’s disease: a Belgian case report
- Diagnostic challenge between a frequent polygenic hypocholesterolemia and an unusual Smith Lemli Opitz syndrome related to bi-allelic DHCR7 mutations
- First reported co-occurrence of Philadelphia chromosome-positive B-cell acute lymphoblastic leukemia with pseudo Chediak-Higashi anomaly and complex karyotype
- Facing the new IVD Regulation 2017/746: Contract Research Organizations (CROs), key partners of IVDs manufacturers for compliance
- Comprehensive analysis and clinical case studies on pseudoeosinophilia: insights and implications – unraveling the complexity: analytical approaches and clinical significance
- Misdiagnosis of type 2B von Willebrand disease as immune thrombocytopenia in a thrombocytopenic patient
- Biological matrices, reagents and turnaround-time: the full-circle of artificial intelligence in the pre-analytical Phase. Comment on Turcic A, et al., Machine learning to optimize cerebrospinal fluid dilution for analysis of MRZH reaction. CCLM 2024;62:436–41
Articles in the same Issue
- Frontmatter
- Editorials
- EFLM European Urinalysis Guideline
- Clinical Chemistry Laboratory Medicine in the post-acute COVID-19 era
- EFLM Guideline
- The EFLM European Urinalysis Guideline 2023
- Review
- Approaching sustainability in Laboratory Medicine
- Opinion Papers
- New reimbursement models to promote better patient outcomes and overall value in laboratory medicine and healthcare
- Screening for sickle cell disease: focus on newborn investigations
- Genetics and Molecular Diagnostics
- The role of Killer immunoglobulin-like receptors (KIRs) in the genetic susceptibility to non-celiac wheat sensitivity (NCWS)
- General Clinical Chemistry and Laboratory Medicine
- Understanding the limitations of your assay using EQA data with serum creatinine as an example
- Add-on testing: stability assessment of 63 biochemical analytes in centrifuged and capped samples stored at 16 °C
- Smartphone swabs as an emerging tool for toxicology testing: a proof-of-concept study in a nightclub
- Allergy: Evaluation of 16 years (2007–2022) results of the shared external quality assessment program in Belgium, Finland, Portugal and The Netherlands
- Cancer Diagnostics
- Monoclonal whole IgG impairs both fibrin and thrombin formation: hemostasis and surface plasmon resonance studies
- Cardiovascular Diseases
- Patients with antiphospholipid syndrome and a first venous or arterial thrombotic event: clinical characteristics, antibody profiles and estimate of the risk of recurrence
- Letters to the Editor
- Familial dysalbuminemic hyperthyroxinemia coexisting with a Grave’s disease: a Belgian case report
- Diagnostic challenge between a frequent polygenic hypocholesterolemia and an unusual Smith Lemli Opitz syndrome related to bi-allelic DHCR7 mutations
- First reported co-occurrence of Philadelphia chromosome-positive B-cell acute lymphoblastic leukemia with pseudo Chediak-Higashi anomaly and complex karyotype
- Facing the new IVD Regulation 2017/746: Contract Research Organizations (CROs), key partners of IVDs manufacturers for compliance
- Comprehensive analysis and clinical case studies on pseudoeosinophilia: insights and implications – unraveling the complexity: analytical approaches and clinical significance
- Misdiagnosis of type 2B von Willebrand disease as immune thrombocytopenia in a thrombocytopenic patient
- Biological matrices, reagents and turnaround-time: the full-circle of artificial intelligence in the pre-analytical Phase. Comment on Turcic A, et al., Machine learning to optimize cerebrospinal fluid dilution for analysis of MRZH reaction. CCLM 2024;62:436–41

