Abstract
Context
Autism spectrum disorder (ASD) is a neurodevelopmental condition characterized by challenges in social communication and repetitive behaviors. Its etiology is influenced by a combination of genetic and environmental factors. Variations in the methylenetetrahydrofolate reductase (MTHFR) gene, which is implicated in folate metabolism and neurodevelopment, are widespread in the autism population. Understanding the relationship between MTHFR gene variations and ASD may be critical for early diagnosis and intervention.
Objectives
This study aims to investigate the association between MTHFR gene variations and the severity of ASD symptoms in a clinical cohort. The goal is to determine whether reduced MTHFR activity correlates with increased symptom severity, thus offering insights into potential mechanisms and intervention strategies.
Methods
A cohort of 78 patients diagnosed with ASD who had previously undergone genetic testing to measure MTHFR activity levels were recruited. ASD severity was assessed utilizing DSM-5 criteria. Statistical analyses were performed to evaluate the relationship between MTHFR activity and ASD symptom severity.
Results
The analysis identified a significant negative correlation between MTHFR activity levels and ASD severity (p<0.05). Patients with lower MTHFR activity exhibited more severe ASD symptoms, as measured by DSM-5 classifications. These findings emphasize the potential link between MTHFR gene variations and neurodevelopmental outcomes in ASD.
Conclusions
This study highlights the role of MTHFR gene variations in modulating ASD severity. The results support the potential for utilizing MTHFR activity as a biomarker for early screening and tailoring targeted interventions for individuals with MTHFR deficiencies. Due to a small sample size, any conclusions drawn from this study are limited and may be misleading in future studies. Further research is warranted to explore the underlying mechanisms and to develop clinical strategies that mitigate the impact of these genetic variations on ASD progression.
Autism spectrum disorder (ASD) is a complex neurodevelopmental condition that presents with a range of social communication deficits and repetitive behaviors. The DSM-5 states that ASD is defined as “persistent deficits in social communication and social interaction across multiple contexts” as well as “restricted, repetitive patterns of behavior, interests, or activities” [1]. Contexts for deficits in social communication include deficits in social-emotional reciprocity, deficits in nonverbal communicative behaviors utilized for social interaction, and deficits in developing, maintaining, and understanding relationships. Restricted, repetitive patterns of behavior, interests, or activities, is manifested by at least two of the following [1]:
Stereotyped or repetitive motor movements, use of objects, or speech.
Insistence on sameness, inflexible adherence to routines, ritualized patterns, and verbal or nonverbal behavior.
Highly restricted, fixated interests that are abnormal in intensity or focus.
Hyper- or hyporeactivity to sensory input or unusual interests in sensory aspects of the environment.
These contexts are not all-encompassing but do well to summarize the definition of ASD. In addition to these two main diagnostic criteria, the DSM-5 also states that symptoms must be present in the early developmental period and cause clinically significant impairment in social, occupational, or other important areas of current functioning. The DSM-5 also states that these disturbances should not be better explained by intellectual disability or global developmental delay. The severity of ASD is primarily based on social communication impairments and restricted repetitive patterns of behavior. ASD severity falls into three different levels of severity based on the degree of support that the individual requires.
The severity of ASD symptoms varies widely among individuals, influenced by a combination of genetic and environmental factors. Despite advances in understanding ASD, research on the genetic basis of symptom severity remains sparse, particularly within the underserved special needs population. However, in this study’s cohort, many patients with ASD receive genetic testing via Genomind, a specialty laboratory certified by the College of American Pathologists (CAP) and the Clinical Laboratory Improvement Amendments (CLIA). Genomind analyzes numerous genes and provides information on the level of activity of said genes. Upon a review of the results of Genomind, a trend was noticed in which patients with severe ASD had low MTHFR gene activity.
MTHFR is an enzyme crucial for folate and homocysteine metabolism. Variations in the MTHFR gene, specifically C677T and A1298C, can reduce enzyme activity, which we theorize may impact the severity of neurodevelopmental diseases [2], [3], [4], [5], [6], [7], [8]. This study investigates whether MTHFR activity levels correlate with the severity of ASD, aiming to enhance our understanding of the genetic factors that influence ASD severity and potentially inform more targeted therapeutic approaches.
Methods
Study design and participants
This study was approved by the Institutional Review Board (IRB) of Cayuse IRB, ensuring compliance with ethical guidelines for research involving human participants (Approval Number: PRO-2022-198, Date: 7/5/2022). This study occurred from July 2023 to September 2023. A retrospective chart review was performed. The review included an analysis of previous psychiatric evaluations, previous diagnoses, and physician notes from previous visits.
Participants included individuals from Rowan’s Regional Integrated Special Needs (RISN) Center diagnosed with ASD who had undergone MTHFR genetic testing through Genomind. Genomind is a precision health company specializing in personalized medicine through pharmacogenetic testing. They integrate genetic, environmental, and lifestyle factors to optimize patient treatment, particularly in mental health. Their solutions aim to enhance medication management by providing healthcare providers with genetic insights to individualize patient care [9]. Genomind classifies MTHFR activity into four levels: low/absent, low to intermediate, intermediate, and high/normal. This is determined through assessing whether a patient has the common mutations of their MTHFR genes: C677T and A1298C on chromosome 1 [2], 10], 11].
Genomind utilizes an SNP panel to determine mutations in the MTHFR gene. A C677T mutation reduces the enzymatic activity of MTHFR by approximately 35 % per T allele, and a A1298C mutation reduces the enzymatic activity of MTHFR by approximately 20 % per C allele [9]. These decreases were categorized by Genomind into the four levels described previously. Calculation of the percent of MTHFR activity decrease leads to category 4 having a 0–20 % decrease, category 3 having a 21–35 % decrease, category 2 having a 36–55 % decrease, and a category 1 having a 56–70 % decrease.
Assessment of MTHFR and ASD severity
MTHFR levels were assigned numeric values from 1 to 4 in conjunction with the previous Genomind classification [9]:
Low/absent
Low to intermediate
Intermediate
High/normal
ASD severity was assessed utilizing DSM-5 criteria [1], with the levels defined as follows:
Level 1=Requires little support
Level 2=Requires substantial support
Level 3=Requires very substantial support
Each patient was given an ASD severity level according to the qualifications from the DSM-5. ASD severity assessments were made by two medical students and confirmed by a treating psychologist (W.A.) and physician (A.I.), based on comprehensive reviews of medical and psychiatric records. Initially, there were 215 subjects in the study from the RISN Center that had Genomind studies performed. Thirty-one patients with MTHFR deficiencies but without an ASD diagnosis were excluded from the analysis, and 106 patients with an ASD diagnosis but without a Genomind analysis were also excluded from the analysis. The number of participants in the study was 78 once the patients were selected for having both an ASD diagnosis and had MTHFR testing performed.
Data analysis
Data analysis involved performing statistical comparisons between ASD severity levels utilizing analysis of variance (ANOVA), with significance defined at p<0.05. The ANOVA utilized the MTHFR levels assigned through Genomind for each patient in a specific ASD category. The average age in years of subjects was also calculated for each ASD severity category along with the age range, number of males, and number females.
Results
The demographics of the 78 participants are as follows:
In the ASD level 1 group, the age range was from 13 to 63 years old with an average age of 26 years old. The number of males was 17, and the number of females was 9.
In the ASD level 2 group, the age range was from 10 to 65 years old with an average age of 29 years old. The number of males was 17, and the number of females was 8.
In the ASD level 3 group, the age range was from 16 to 60 years old with an average age of 25 years old. The number of males was 17, and the number of females was 10.
The ASD severity, average MTHFR levels, and number of subjects from the 78 patients are included in Table 1.
Summarized ASD and MTHFR level data from 78 subjects.
| ASD level severity | Number of subjects | Average MTHFR level | Age range, years | Average age, years | Number of males | Number of females |
|---|---|---|---|---|---|---|
| 1 | 26 | 2.91 | 13–63 | 26 | 17 | 9 |
| 2 | 25 | 2.32 | 10–65 | 29 | 17 | 8 |
| 3 | 27 | 2.03 | 16–60 | 25 | 17 | 10 |
-
ASD, autism spectrum disorder; MTHFR, methylenetetrahydrofolate reductase.
Analysis of the data demonstrated a negative correlation between MTHFR activity levels and ASD severity. Patients with lower MTHFR activity were more likely to have higher ASD severity levels. ANOVA revealed significant differences between ASD levels, demonstrating a p value of 0.0048. This may indicate a statistically significant relationship between reduced MTHFR activity and increased ASD severity. Average MTHFR levels in the different ASD levels are summarized in Figure 1 below.
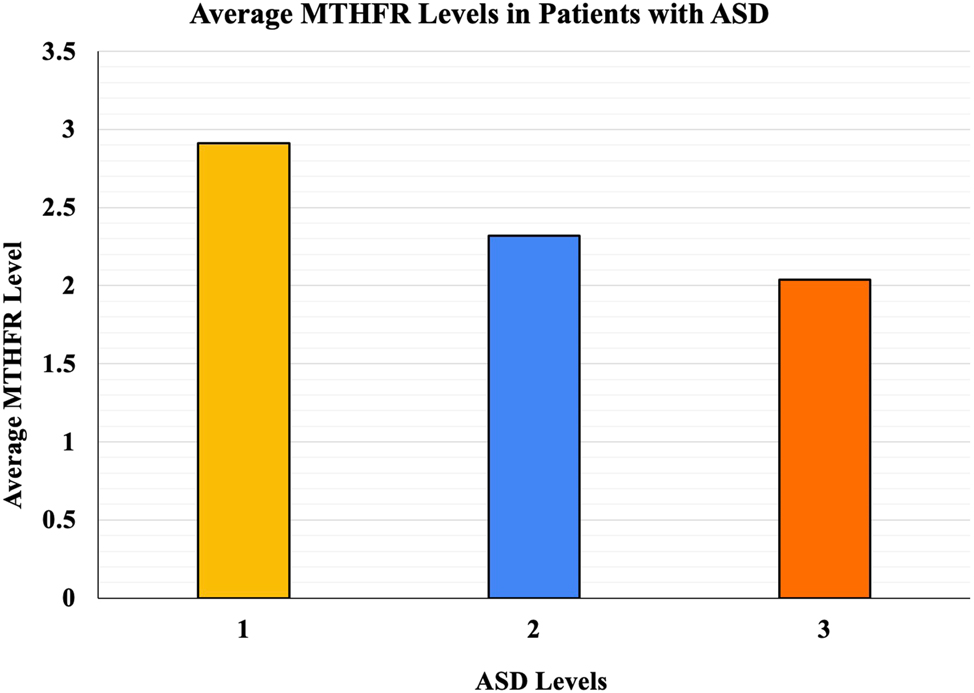
The average MTHFR levels in patients with ASD.
Discussion
Our findings suggest that lower MTHFR activity is associated with an increased severity of ASD, supporting the role of genetic factors in influencing the condition. These results align with previous studies highlighting the importance of MTHFR in neurodevelopment and its potential impact on ASD symptomatology.
Contextualizing findings
The observed correlation between reduced MTHFR activity and ASD severity adds to the growing body of evidence emphasizing the role of metabolic and genetic pathways in neurodevelopmental disorders. While our findings corroborate prior research, they also provide a more specific understanding of the clinical implications of MTHFR activity in ASD severity. For example, our study highlights not only the existence of this correlation, but also the potential for utilizing MTHFR activity levels as a biomarker to stratify risk and guide interventions.
Clinical applications
These insights are clinically significant, because they pave the way for personalized interventions. We theorize that screening for MTHFR variants at the time of ASD diagnosis or even during infancy could allow for earlier interventions to mitigate symptom severity. Early treatment strategies might include targeted nutritional support or other therapies to address MTHFR deficiencies, potentially improving long-term outcomes for individuals with ASD. Such an approach would be especially valuable for osteopathic physicians, who often emphasize holistic and preventative care.
Comparison with previous studies
Our findings are consistent with earlier studies that have linked folate metabolism and neurodevelopment, but they differ in providing quantitative evidence of how MTHFR activity correlates with ASD severity. Unlike prior research that primarily explored MTHFR variants qualitatively, our study quantifies their impact, offering a new dimension to understanding their clinical relevance.
Limitations
Several limitations of this study must be acknowledged. First, the sample size of 78 participants, while sufficient to detect significant correlations, limits the generalizability of our findings. This small sample size also means that any conclusions drawn from this study are limited and may be misleading in future studies. Second, the cross-sectional design of the study precludes us from drawing causal conclusions about the relationship between MTHFR activity and ASD severity. Third, potential confounding factors, such as environmental influences and comorbid conditions, were not fully accounted for in our analysis. This is mainly due to the complex nature of the confounding factors and the difficulty quantifying their impact on the data. Future longitudinal studies with larger and more diverse cohorts are needed to validate our findings and explore these additional variables.
Conclusions
The findings of this study suggest that reduced MTHFR activity is associated with increased severity of ASD, highlighting the potential role of genetic factors in the manifestation and progression of ASD symptoms. This association underscores the importance of considering genetic profiles, such as MTHFR variations, in understanding the complex etiology of ASD. These results support the possible benefits of incorporating genetic testing for MTHFR activity levels into clinical practice, especially during the early stages of ASD diagnosis. Early identification of individuals with lower MTHFR activity could enable more personalized and targeted interventions, such as nutritional and pharmacological strategies aimed at mitigating the effects of MTHFR deficiencies. Furthermore, these findings open avenues for additional research into the broader neurobehavioral consequences of MTHFR deficiencies beyond the ASD population. By expanding our understanding of how MTHFR variations influence neurodevelopment, this research could contribute to more effective and individualized treatment strategies, ultimately improving outcomes and quality of life for individuals with ASD.
-
Research ethics: This study was approved by the Institutional Review Board (IRB) of Cayuse IRB, ensuring compliance with ethical guidelines for research for research involving human participants (approval number: PRO-2022-198).
-
Informed consent: Not applicable.
-
Author contributions: The authors have accepted responsibility for the entire content of this manuscript and approved its submission.
-
Use of Large Language Models, AI and Machine Learning Tools: None declared.
-
Conflict of interest: None declared.
-
Research funding: None declared.
-
Data availability: All raw data may be obtained on request from the corresponding author.
References
1. American Psychiatric Association. Diagnostic and statistical manual of mental disorders, 5th ed. Washington, D.C.: American Psychiatric Association; 2013.10.1176/appi.books.9780890425596Search in Google Scholar
2. El-Baz, F, El-Aal, MA, Kamal, TM, Sadek, AA, Othman, AA. Study of the C677T and 1298AC polymorphic genotypes of MTHFR gene in autism spectrum disorder. Electron Physician 2017;9:5287–93. https://doi.org/10.19082/5287.Search in Google Scholar PubMed PubMed Central
3. Fang, Y, Cui, Y, Yin, Z, Hou, M, Guo, P, Wang, H, et al.. Comprehensive systematic review and meta-analysis of the association between common genetic variants and autism spectrum disorder. Gene 2023;887:147723. https://doi.org/10.1016/j.gene.2023.147723.Search in Google Scholar PubMed
4. Hoxha, B, Hoxha, M, Domi, E, Gervasoni, J, Persichilli, S, Malaj, V, et al.. Folic acid and autism: a systematic review of the current state of knowledge. Cells 2021;10:1976. https://doi.org/10.3390/cells10081976.Search in Google Scholar PubMed PubMed Central
5. Li, Y, Qiu, S, Shi, J, Guo, Y, Li, Z, Cheng, Y, et al.. Association between MTHFR C677T/A1298C and susceptibility to autism spectrum disorders: a meta-analysis. BMC Pediatr 2020;20:449. https://doi.org/10.1186/s12887-020-02330-3.Search in Google Scholar PubMed PubMed Central
6. Roufael, M, Bitar, T, Sacre, Y, Andres, C, Hleihel, W. Folate-methionine cycle disruptions in ASD patients and possible interventions: a systematic review. Genes (Basel) 2023;14:709. https://doi.org/10.3390/genes14030709.Search in Google Scholar PubMed PubMed Central
7. Xu, S, Men, S, Wang, X, Zhan, F, Yuan, X. Association of MTHFR gene C677T polymorphism with problem behavior and inheritance pattern among children with autism. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2022;39:898–902 [Chinese]. https://doi.org/10.3760/cma.j.cn511374-20201224-00905.Search in Google Scholar PubMed
8. Qiu, S, Qiu, Y, Li, Y, Cong, X. Genetics of autism spectrum disorder: an umbrella review of systematic reviews and meta-analyses. Transl Psychiatry 2022;12:249. https://doi.org/10.1038/s41398-022-02009-6.Search in Google Scholar PubMed PubMed Central
9. Genomind. The science behind genomind. Genomind. Available from: https://genomind.com/the-science/ [Accessed 29 Dec 2024].Search in Google Scholar
10. Ismail, S, Senna, AA, Behiry, EG, Ashaat, EA, Zaki, MS, Ashaat, NA, et al.. Study of C677T variant of methylene tetrahydrofolate reductase gene in autistic spectrum disorder Egyptian children. Am J Med Genet B Neuropsychiatr Genet 2019;180:305–9. https://doi.org/10.1002/ajmg.b.32729.Search in Google Scholar PubMed
11. Wei, H, Zhu, Y, Wang, T, Zhang, X, Zhang, K, Zhang, Z. Genetic risk factors for autism-spectrum disorders: a systematic review based on systematic reviews and meta-analysis. J Neural Transm (Vienna) 2021;128:717–34. https://doi.org/10.1007/s00702-021-02360-w.Search in Google Scholar PubMed
© 2025 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.

