Abstract
Objectives
There are several hypotheses on the effects of the rs1738074 T/C single nucleotide polymorphism in the TAGAP gene; however, there has been no study on Turkish pediatric patients. We aimed to investigate the association of celiac disease (CD) and type 1 diabetes mellitus (T1DM) comorbidity with the polymorphism in the TAGAP gene of Turkish pediatric patients.
Methods
Totally, 127 pediatric CD patients and 100 healthy children were included. We determined the polymorphism by the allele-specific polymerase chain reaction method. We used IBM SPSS Statistics version 25.0 and Arlequin 3.5.2 for the statistical analyses. The authors have no conflict of interest.
Results
It was determined that 72% (n=154) of only CD patients had C allele, whereas 28% (n=60) had T allele. Of the patients with celiac and T1DM, 42.5% (n=17) and 57.5% (n=23) had T and C alleles, respectively. Of the individuals in control group, 67% (n=134) had C allele, whereas 33% (n=66) had T allele.
Conclusions
There was no significant difference in the genotype and allele frequencies between the patient and control groups (p>0.05). There was no significant association between the disease risk and the polymorphism in our study group.
Öz
Amaç
Literatürde TAGAP genindeki rs1738074 T/C tek nükleotit polimorfizminin (SNP) hastalıklarla ilişkisine yönelik birçok çalışma bulunmaktadır, ancak Türk pediyatrik hastalarda rs1738074 T/C polimorfizmi ile ilgili bir çalışma mevcut değildir. Bu çalışmada, Türk pediyatrik hastalarda çölyak ve tip 1 diyabet komorbiditesinin TAGAP genindeki polimorfizmle ilişkisinin araştırılması amaçlandı.
Gereç ve Yöntem
Çalışmaya 127 çölyak hastası ve 100 sağlıklı çocuk dahil edildi. Polimorfizmi saptamak için allel spesifik polimeraz zincir reaksiyonu yöntemi kullanıldı. IBM SPSS Statics version 25.0 ve Arlequin 3.5.2 programları kullanılarak istatistiksel analizler gerçekleştirildi. Yazarların herhangi bir çıkar çatışması bulunmamaktadır.
Bulgular
Sadece Çölyak hastalarının %72’si (n=154) C alleline sahipken, %28’inin (n=60) T alleline sahip olduğu belirlendi. Çölyak ve tip 1 diyabetli hastaların %42.5’i (n=17) T alleline, %57.5’i (n=23) C alleline sahipti. Kontrol grubundaki bireylerin %67’sinin (n=134) C alleline, %33’ünün (n=66) ise T alleline sahip olduğu gözlendi.
Sonuç
Hasta ve kontrol grubu arasında genotip ve allel frekansları bakımından anlamlı bir farklılık yoktur (p>0.05). Çalışma grubumuzda hastalık riskiyle rs1738074 T/C polimorfizmi arasında anlamlı bir ilişki bulunamamıştır.
Introduction
Celiac disease (CD) is a systemic enteropathy of the small intestine of which development and symptoms are affected by environmental factors and genetic predisposition [1], [, 2]. The best-known environmental factor is gluten that is the name of the proteins found in wheat, barley, rye, and cereal [2]. Different gluten peptides can activate the adaptive or innate immune response in CD [3].
The primary genetic factor of CD is the HLA DQ allele. Most of the celiac patients have HLA DQB1*02 and DQA1*05 (HLA DQ2.5) allotype that presents gluten peptides to gluten-reactive CD4+ T cells in CD. It infrequently develops in the lack of HLA DQ2.5 heterodimers, and that the predisposing HLA DQ2.5 subtypes are necessary, but not competent for causing the CD [4]. Furthermore, the investigators have previously shown an association between CD and other autoimmune diseases. Type 1 diabetes mellitus (T1DM) is one of these autoimmune disorders, and almost 5% of CD patients have T1DM [5]. rs1738074 T/C single nucleotide polymorphism (SNP) has been found associated with CD and T1DM (p=1.7 x 10−4) [6].
CD is diagnosed by intestine biopsy [7]. Detection of the IgA class serum specific antibodies against transglutaminase protein is also a part of the diagnosis [8]. Besides, the experts can use the results of IgA antibody screening and other serological methods together to diagnose CD [7]. Numerous researchers have attempted to find different detection methods and biomarkers to diagnose CD without invasive procedures. Many more risk loci outside the HLA region have been identified as disease biomarkers. Recently, genome-wide association studies have identified several possible inherited risk factors for CD, such as T-cell activation Rho GTPase activating protein (TAGAP) [9].
TAGAP encodes a T cell activation Rho GTPase activating protein important in regulating cytoskeletal construction in the T cell activation. This protein has a vital role in the regulation of T cell-driven autoimmune diseases [9], [, 10]. In the Rho GTPase pathway, inactive GDP-bound state changes to an active GTP-bound state. This reaction is catalyzed by GDP exchange factors (GEFs). GEF activated Rho interacts with ROCK to trigger the proteins, which play an essential role in focal adhesion, cytoskeletal rearrangement, and stabilization of the actin cytoskeleton. TAGAP negatively regulates the pathway, and therefore, causes the dysfunctional actin protein rearrangement [11].
rs1738074 T/C polymorphism of the TAGAP gene and several autoimmune diseases have been studied in different populations [10], [12], [13]. The association between TAGAP-multiple sclerosis and TAGAP-CD were investigated in Iranian and in the Swedish-Norwegian community, respectively [12], [, 13]. In this study, we aimed to evaluate the association of TAGAP rs1738074 polymorphism with the development of CD in the Turkish pediatric population. Also, TAGAP polymorphism was assessed in cases with celiac and T1DM comorbidity. HLA DQB1/DQA1 allele frequencies and tissue transglutaminase (tTGA) levels of the patients were also included in interpreting the results.
Materials and methods
Study population
In this study cohort, 127 pediatric patients with CD and 100 healthy children were screened for TAGAP rs1738074 T/C SNP [14]. The DNA samples were used retrospectively for the allele-specific PCR method to determine genotype frequency. Of the children, 107 (84.3%) were diagnosed with only CD, and 20 (15.7%) were diagnosed with diabetes and CD. CD was diagnosed according to the ESPGHAN criteria in the cases [15].
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1975 Helsinki declaration and its later amendments or comparable ethical standards. Written consents of the volunteers were obtained.
DNA isolation
The DNA samples were isolated using the EZ1 DNA Blood isolation Kit (Qiagen, USA) on Geno M-6 Genevision DNA isolation instrument. The procedure was performed according to the manufacturer’s instructions. 50 µL DNA was obtained from each 200 µL blood sample with EDTA. The purity and concentration of DNA samples were measured by Nanodrop 2000 Spectrophotometer (Thermo Scientific, USA). The concentrations of the DNA samples were above 20 ng/μL, and their purities were between 1.75 and 1.85.
TAGAP polymorphism by allele-specific PCR method
Allele-specific PCR was used to determine genotype and allele frequencies. This method consists of two steps: PCR amplification and agarose gel electrophoresis. Four primers were synthesized using previous primer regions (Oligomer Biotechnology, Turkey) [12]:
Reverse T (allele T): 5′-GCTGTAAAGAATGGGAGAAACAGaAt-3′
Reverse C (allele C): 5′-GCTGTAAAGAATGGGAGAAACAGaAc-3′
Forward outer: 5′-GCCCTAAAAGGAATGAGGAAGC-3′
Reverse outer: 5′-GAGCATATTCCACGGGATAGC-3′
According to this method, PCR reactions were carried out using the four primers in Eppendorf tubes containing 1 µL MgCl2 (50 mM), 1 µL dNTP (10 mM), 2.5 µL 10× PCR buffer, 0.25 µL Taq DNA Polymerase (5 unit/L), 1 µL DNA (100 ng/μL), 1 µL reverse C (10 pM), 1 µL reverse T (10 pM), 0.25 µL reverse outer primer (10 pM), 0.50 µL forward outer primer (10 pM), and deionized H2O up to 25 µL. The PCR conditions were as 5 min denaturing step at 94 °C, 40 s denaturing step at 94 °C, 45 s annealing step at 60 °C, and extension step at 72 °C for 50 s, for a total of 31 cycles, and 10 min at 72 °C. Subsequently, we analyzed the amplicons under the UV light after separating them on 2% agarose gel electrophoresis [12].
HLA-DQB1/DQA1 molecular typing
HLA-DQB1/DQA1 typing was performed by sequence-specific oligonucleotide probe method. This method includes two steps: Amplification and hybridization. According to the manufacturer’s instructions (Lifecodes HLA SSO Typing Kit Immucor, USA), we added 10 µL sterile distilled water, 6 µL mastermix, 4 µL DNA sample, and 0.2 µL Taq DNA polymerase into the 0.2 mL Eppendorf tube for each patient. The tubes were placed in thermal cycler instrument. The amplification PCR conditions were: 3 min at 95 °C for one cycle, 12 cycles of 15 s at 95 °C, 30 s at 60 °C, 30 s at 72 °C, 28 cycles of 10 s at 95 °C, 30 s at 63 °C, 30 s at 72 °C, one cycle of 2 min at 72 °C, and holding at 4 °C.
For the second step (hybridization step), we put 5 µl amplicon into a well in the 96 well plate. We added 15 µL-preheated beads on the sample. The hybridization PCR conditions were 2 min at 97 °C, 10 min at 47 °C, 8 min at 56 °C, and holding at 56 °C.
After polymerase chain reaction, 0.85 µL streptavidin was diluted in 170 µL dilution solution for each well, and we added the diluted streptavidin on the sample at 56 °C. We used Luminex xMAP technology to analyze the samples. The results were evaluated by MatchIT Software Program.
Statistical analyses
Statistical analyses were performed using IBM SPSS Statistics version 25.0. We determined the allele frequencies by descriptive statistics. A chi-squared test (χ2) was performed to determine the association between allele frequency and CD. Hardy–Weinberg equilibrium test was performed to analyze the consistency of genotype frequencies with the equilibrium. HLA DQ2.5 allele frequencies were calculated by Arlequin 3.5.2.
Results
In this study, 127 pediatric patients (107 celiac patients [84.3%] and 20 [15.7%] celiac + diabetes patients) and 100 controls were compared according to their TAGAP rs1738074 T/C SNP and HLA DQ2.5 alleles to evaluate their association with the CD. The mean ages of the patient and control groups were 8.7 ± 4.8 and 9.9 ± 5.9, respectively. Of the patients, 59% (n=75) were female, whereas 41% (n=52) were male. Of the healthy individuals, 46% (n=46) were female, whereas 54% (n=54) were male.
As presented in Figure 1, the 372 bp (the forward outer and reverse outer primers) band shows the internal PCR control confirming the polymerase chain reaction in the PCR tubes. The 224 bp (the forward outer and reverse C/T primers) band shows the specific band.
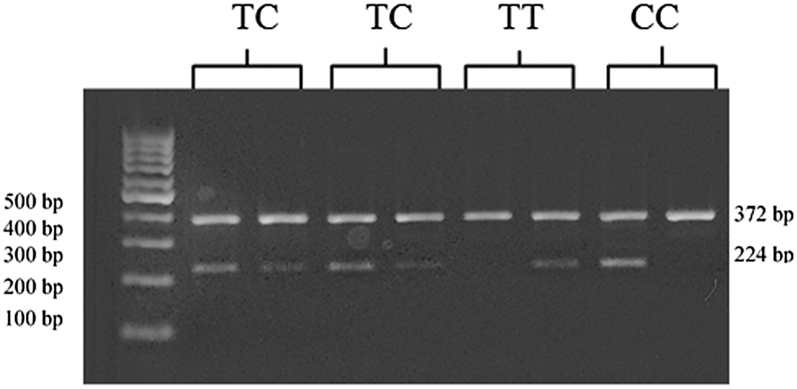
Agarose gel electrophoresis of the genotypes. Lane 2, 3, 4, and 5 show heterozygous TC individuals. Lanes 6–7 and 8–9 shows homozygous individuals for TT and CC genotypes, respectively. The DNA size marker is in lane 1
Analysis of the TAGAP polymorphism revealed a higher level of the CC genotype and C allele in the all patients than the control group. There was no significant difference in the genotype and allele frequencies between the patient and the control groups (Table 3). The association between the age of the diagnosis of CD and genotypes and alleles was evaluated; however, no statistically significant result has been achieved.
The genotype frequencies of the two groups were compatible with Hardy–Weinberg equilibrium (patient group p>0.05 and control group p>0.05) (Table 1). The frequencies of TT, TC, and CC genotypes were not significantly different between celiac and celiac + diabetes patient groups (p>0.05).
Comparison of the TAGAP genotype and allele frequencies between patient and control groups.
| Genotype frequencies | Allele frequencies | ||||
|---|---|---|---|---|---|
| TT | CC | TC | T allele | C allele | |
| Control (n=100) | 16% (n=16) | 50% (n=50) | 34% (n=34) | 33% (n=66) | 67% (n=134) |
| Celiac patient (n=107) | 14.9% (n=16) | 58.9% (n=63) | 26.1% (n=28) | 28% (n=60) | 72% (n=154) |
| Celiac + T1DM (n=20) | 20% (n=4) | 35% (n=7) | 45% (n=9) | 42.5% (n=17) | 57.5% (n=23) |
| P | >0.05 | >0.05 | |||
T1DM, type 1 diabetes mellitus
We also analyzed the frequencies of HLA DQB1 and HLA DQA1 alleles in the celiac and control groups (Table 2). The frequency of HLA DQ2.5 alleles in patients was significantly higher than the control group (p<0.001).
The frequency of HLA DQB1 and HLA DQA1 alleles in celiac patients and controls.
| The ratios of patient alleles (n=254) | The ratios of the control alleles (n=200) | p<0.001 | |
| DQB1* | |||
| 02 | 50% (n=127) | 11.5% (n=23) | |
| 03 | 28.7% (n=73) | 42.5% (n=85) | |
| 04 | 0.4% (n=1) | 2% (n=4) | |
| 05 | 14.6% (n=37) | 22.5% (n=45) | |
| 06 | 6.3% (n=16) | 21.5% (n=43) | |
| DQA1* | p<0.001 | ||
| 01 | 16.3% (n=41) | 41.5% (n=83) | |
| 02 | 17.9% (n=45) | 4.5% (n=9) | |
| 03 | 15.1% (n=38) | 20.5% (n=41) | |
| 04 | 0.4% (n=1) | 1% (n=2) | |
| 05 | 49.2% (n=125) | 31% (n=62) | |
| 06 | 0.4% (n=1) | 1.5% (n=3) | |
| 10 | 0.4% (n=1) | n=0 | |
The most of celiac patients carry HLA DQ2.5 heterodimers, encoded by HLA DQ2.5 alleles. Therefore, TAGAP rs1738074 genotype and allele frequencies were compared in patients and controls with and without HLA DQ2.5 allele frequencies. There was no statistically significant association between the groups (p>0.05). The most frequently observed genotype was CC in both groups (Table 3).
Comparison of genotype and allele frequencies between controls, patient with HLA DQ2.5 alleles, patients except HLA DQ2.5 alleles.
| TT | CC | TC | T | C | |
|---|---|---|---|---|---|
| HLA DQ2.5 control (n=16) | 12.5% (n=2) | 62.5% (n=10) | 25% (n=4) | 33% (n=8) | 67% (n=24) |
| HLA DQ2.5 patients (n=85) | 18.8% (n=16) | 51.8% (n=44) | 29.4% (n=25) | 33.5% (n=57) | 66.5% (n=113) |
| p | >0.05 | >0.05 | |||
| HLA alleles except HLA DQ2.5 control (n=84) | 17.9% (n=15) | 47.6% (n=40) | 34.5% (n=29) | 35% (n=59) | 65% (n=109) |
| HLA alleles except HLA DQ2.5 (n=42) | 9.5% (n=4) | 59.5% (n=25) | 31% (n=13) | 25% (n=21) | 75% (n=63) |
| p | >0.05 | >0.05 | |||
It was reported that strong positive tTGA levels (≥100 U/A) were related to some symptoms of CD [16]. We compared the TAGAP rs1738074 genotype and allele frequencies of the patients with this enzyme activity. There was no significant correlation between the tTGA level and the polymorphisms (Table 4). Besides, we investigated the association of HLA DQ2.5 alleles with tTGA activity. The results were not statistically significant (p>0.05), although we observed higher activity in the HLA DQ2.5 patients than the other patients.
Comparison of genotype and allele frequencies between individuals with enzyme specific antibody level (tTGA).
| TT | CC | TC | T | C | |
|---|---|---|---|---|---|
| tTGA<100 (n=22) | 9.1% (n=2) | 59.1% (n=13) | 31.8% (n=7) | 25% (n=11) | 75% (n=33) |
| tTGA>100 (n=105) | 17.2% (n=18) | 53.3% (n=56) | 29.5% (n=31) | 32% (n=67) | 68% (n=143) |
| p | >0.05 | >0.05 | |||
tTGA, tissue transglutaminase antibody.
Discussion
CD is an autoimmune disease characterized by the histopathological abnormalities of the small intestine [2], [, 17]. It has been considered that high risk of the disease was related to specific HLA DQ genotypes [1], [, 18]. HLA DQ group is one of the HLA class II molecules that present peptides to T cells, and it consists of polymorphic alpha and beta chains of the DQ molecule [19], [, 20]. Association of HLA alleles with CD were studied in adults, and only 40% of the CD pathogenesis was found correlated with HLA DQ2.5 molecules [21]. Different loci can be a risk factor for CD development, although the patient with these HLA alleles had CD. Therefore, HLA cannot be the only reason for the CD [9]. In our study, we noticed a higher frequency of DQ2.5 in the patient group than the control group.
In recent years, the risk of CD has been associated with polymorphisms in some other non-HLA genetic regions, such as the TAGAP region. TAGAP gene encodes a Rho GTPase that plays a role in the regulation of the cytoskeletal modifications and the association of this gene with autoimmune diseases including rheumatoid arthritis, multiple sclerosis, and CD has been reported previously [18]. The studies showed that this protein could be a negative regulator of Rho-GTPase signaling in CD. In a meta-analysis study, Huang et al. [9] showed that TAGAP rs1738074 T/C polymorphism might have effect on developing CD due to the regulation of the Rho-GTPase cycle. The patients with this polymorphism can have CD compared to the patients who do not carry this polymorphism. Their meta-analysis showed a statistically significant correlation between CD and TAGAP rs1738074 T/C polymorphism [9], [, 12]. Another study included many American and European populations (4,533 patients and 10.750 controls) and confirmed that the polymorphism was significantly associated with the CD susceptibility [22]. However, the results were not statistically significant in our study cohorts. To our knowledge, this is the first study investigating the association of TAGAP rs1738074 T/C polymorphism with CD in the Turkish pediatric population.
We analyzed the association of this polymorphism with disease type (only celiac, celiac + T1DM). TAGAP gene previously associated with T1DM. Many non-HLA loci overlap with T1DM and other autoimmune disorders, such as T-cell activation Rho GTPase activating protein (TAGAP) [9]. However, the frequencies of TT, TC, and CC genotypes were not significant between celiac and celiac with diabetes patients.
In a recent study focusing on the celiac siblings, it was suggested that HLA DQ2.5 alleles and TAGAP rs1738074 T/C polymorphism were together more associated with CD [1]. However, we have not encountered any literature investigating the association between TAGAP rs1738074 T/C polymorphism and HLA DQ2.5 alleles. In our pediatric study cohort, the results were not statistically significant.
tTGA positivity usually indicates the earliest stage of autoimmunity, and it has a very high predictive value for CD [18]. For this purpose, we investigated the association of this positivity with TAGAP rs1738074 T/C polymorphism and HLA typing of the patients. There was no significant association between tTGA positivity and the rs1738074 polymorphism. In a large-scale study, the investigators suggested that TAGAP polymorphism was associated with tTGA positivity in pediatric celiac patients [18]. However, their variant (TAGAP rs1054091) was different from our variant (rs1738074). Thus, our study is the first study in which the relation between tTGA positivity and TAGAP rs1738074 T/C polymorphism was analyzed. We also examined the association of HLA typing with tTGA positivity. We discovered that tTGA positivity was higher in HLA DQ2.5 patients. However, our results were not correlated with TAGAP rs1738074 polymorphism.
This is the first study investigating the association of TAGAP rs1738074 polymorphism (T/C) with CD disease and related features in the Turkish pediatric population. Our results indicated that there were no significant associations between the parameters. Although it is stated that CD development is multifactorial and there are many genes affecting its pathogenesis, the molecular determinants underlying the pathogenesis of the CD remain unclear. Therefore, target molecular biomarkers should be investigated to diagnose patients without a biopsy. Further investigations should be conducted, including a large-scale population and other non-HLA gene polymorphisms. In addition, we considered that the relation between tTGA positivity and different TAGAP polymorphism variants might affect disease development. Thus, it is necessary to focus on the relationship between tTGA and different TAGAP polymorphism variants.
Research funding: None declared.
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
Competing interests: Authors state no conflict of interest.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1975 Helsinki declaration and its later amendments or comparable ethical standards. Ethical approval was waived by the local non-interventional Ethics Committee (2019/14-10).
References
1. Izzo, V, Pinelli, M, Tinto, N, Esposito, MV, Cola, A, Sperandeo, MP, et al.. Improving the estimation of celiac disease sibling risk by non-HLA genes. PLoS One 2011;6:e26920. https://doi.org/10.1371/journal.pone.0026920.Search in Google Scholar
2. Lebwohl, B, Sanders, DS, Green, PH. Coeliac disease. Lancet 2018;391:70–81. https://doi.org/10.1016/s0140-6736(17)31796-8.Search in Google Scholar
3. Magni, S, Comani, GB, Elli, L, Vanessi, S, Ballarini, E, Nicolini, G, et al.. miRNAs affect the expression of innate and adaptive immunity proteins in celiac disease. Am J Gastroenterol Suppl 2014;109:1662. https://doi.org/10.1038/ajg.2014.203.Search in Google Scholar
4. Kupfer, SS, Jabri, B. Pathophysiology of celiac disease. Gastrointest Endosc Clin N Am 2012;22:639–60. https://doi.org/10.1016/j.giec.2012.07.003.Search in Google Scholar
5. Salazar, C, García-Cárdenas, JM, Paz-y-Miño, C. Understanding Celiac disease from genetics to the future diagnostic strategies. Clin Med Insights Gastroenterol 2017;10:1–13. https://doi.org/10.1177/1179552217712249.Search in Google Scholar
6. Chen, R, Stahl, EA, Kurreeman, FAS, Grefersen, PK, Siminovitch, KA, Worthington, J, et al.. Fine mapping the TAGAP risk locus in rheumatoid arthritis. Genes Immun 2011;12:314. https://doi.org/10.1038/gene.2011.8.Search in Google Scholar
7. Murray, JA, Frey, MR, Oliva-Hemker, M. Celiac disease. Gastroenterology 2018;154:2005–8. https://doi.org/10.1053/j.gastro.2017.12.026.Search in Google Scholar
8. Franceschini, E, Lionetti, ME, D’Adamo, G, D’Angelo, E, Gatti, S, Catassi, GN, et al.. Misuse of serological screening tests for celiac disease in children: a prospective study in Italy. Dig Liver Dis 2019;51:1547–50. https://doi.org/10.1016/j.dld.2019.06.016.Search in Google Scholar
9. Huang, SQ, Zhang, N, Zhou, ZX, Huang, CC, Zeng, CL, Xiao, D, et al.. Association of LPP and TAGAP polymorphisms with celiac disease risk: a meta-analysis. Int J Environ Res Public Health 2017;14:1–17. https://doi.org/10.3390/ijerph14020171.Search in Google Scholar
10. Arshad, M, Bhatti, A, John, P, Jalil, F, Borghese, F, Kawalkowska, JZ, et al.. T cell activation Rho GTPase activating protein (TAGAP) is upregulated in clinical and experimental arthritis. Cytokine 2018;104:130–5. https://doi.org/10.1016/j.cyto.2017.10.002.Search in Google Scholar
11. Ota, T, Maeda, M, Okamoto, M, Tatsuka, M. Positive regulation of Rho GTPase activity by RhoGDIs as a result of their direct interaction with GAPs. BMC Syst Biol 2015;9:1–9. https://doi.org/10.1186/s12918-015-0143-5.Search in Google Scholar
12. Jazaeri, A, Vallian, S. Association of rs1738074 polymorphism of TAGAP gene with susceptibility to multiple sclerosis in the Iranian population. Neurosci Lett 2017;648:66–9. https://doi.org/10.1016/j.neulet.2017.03.041.Search in Google Scholar
13. Amundsen, SS, Rundberg, J, Adamovic, S, Gudjonsdottir, AH, Ascher, H, Ek, J, et al.. Four novel coeliac disease regions replicated in an association study of a Swedish–Norwegian family cohort. Genes Immun 2010;11:79–86. https://doi.org/10.1038/gene.2009.67.Search in Google Scholar
14. National Library of Medicine. Reference SNP report. Available from: https://www.ncbi.nlm.nih.gov/snp/rs1738074 [Accessed May 2020].Search in Google Scholar
15. Husby, S, Koletzko, S, Korponay-Szabo, IR, Mearin, ML, Phillips, A, Shamir, R, et al.. European society for paediatric gastroenterology, hepatology and nutrition guidelines for the diagnosis of coeliac disease. JPGN 2012;54:136–60. https://doi.org/10.1097/mpg.0b013e31821a23d0.Search in Google Scholar
16. Doğan, G, Ayhan, S, Yılmaz, B, Appak, YC, Dundar, PE, Ecemis, T, et al.. Relationship between duodenal histopathology and strong positive tissue transglutaminase antibodies in children with celiac disease. J Curr Pediatr 2015;13:171–6.10.4274/jcp.81994Search in Google Scholar
17. Walker, MM, Ludvigsson, JF, Sanders, DS. Coeliac disease: review of diagnosis and management. Med J Aust 2017;207:173–8. https://doi.org/10.5694/mja16.00788.Search in Google Scholar
18. Sharma, A, Liu, X, Hadley, D, Hagopian, W, Liu, E, Chen, WM, et al.. Identification of non-HLA genes associated with celiac disease and country-specific differences in a large, international pediatric cohort. PLoS One 2016;11:e0152476. https://doi.org/10.1371/journal.pone.0152476.Search in Google Scholar
19. Manczinger, M, Kemény, L. Peptide presentation by HLA-DQ molecules is associated with the development of immune tolerance. PeerJ 2018;6:e5118. https://doi.org/10.7717/peerj.5118.Search in Google Scholar
20. Helmuth, R, Fildes, N, Blake, E, Luce, MC, Chimera, J, Madej, R, et al.. HLA-DQα allele and genotype frequencies in various human populations determined by using enzymatic amplification and oligonucleotide probes. Am J Hum Genet 1990;47:515–23.Search in Google Scholar
21. Sollid, LM. Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol 2002;2:647–55. https://doi.org/10.1038/nri885.Search in Google Scholar
22. Dubois, PC, Trynka, G, Franke, L, Hunt, KA, Romanos, J, Curtotti, A, et al.. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 2010;42:295–302. https://doi.org/10.1038/ng.543.Search in Google Scholar
© 2021 Melek Pehlivan et al., published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Review Articles
- Medicine in philately: History of Quarantine
- Prevention of extra-analytical phase errors by non-analytical automation in clinical laboratory
- Research Articles
- Serum microRNA signature is capable of predictive and prognostic factor for SARS-COV-2 virulence
- The method comparison and the verification of precision of Mindray CL-6000i thyroid function tests (TFTs)
- Measurement of serum creatinine levels with liquid chromatography-tandem mass spectrometry: comparison with Jaffe and enzymatic methods
- The importance of sPD-1, sOX40L and sGITR in terms of clinicopathology and histopathology in gastric cancer
- The cytotoxic and apoptotic effects of Ferulago W. Koch extracts on various cancer cell lines
- Investigation of TAGAP gene polymorphism (rs1738074) in Turkish pediatric celiac patients
- Suberoylanilide hydroxamic acid inhibits LX2 cells proliferation via decreasing yes-associated protein/transcriptional coactivator with PDZ-binding motif proteins
- Effect of protocatechuic acid against renal ischemia reperfusion damage on extracellular matrix integrity and related signal pathways
- The role of zonulin in the pathogenesis of diabetic retinopathy
Articles in the same Issue
- Frontmatter
- Review Articles
- Medicine in philately: History of Quarantine
- Prevention of extra-analytical phase errors by non-analytical automation in clinical laboratory
- Research Articles
- Serum microRNA signature is capable of predictive and prognostic factor for SARS-COV-2 virulence
- The method comparison and the verification of precision of Mindray CL-6000i thyroid function tests (TFTs)
- Measurement of serum creatinine levels with liquid chromatography-tandem mass spectrometry: comparison with Jaffe and enzymatic methods
- The importance of sPD-1, sOX40L and sGITR in terms of clinicopathology and histopathology in gastric cancer
- The cytotoxic and apoptotic effects of Ferulago W. Koch extracts on various cancer cell lines
- Investigation of TAGAP gene polymorphism (rs1738074) in Turkish pediatric celiac patients
- Suberoylanilide hydroxamic acid inhibits LX2 cells proliferation via decreasing yes-associated protein/transcriptional coactivator with PDZ-binding motif proteins
- Effect of protocatechuic acid against renal ischemia reperfusion damage on extracellular matrix integrity and related signal pathways
- The role of zonulin in the pathogenesis of diabetic retinopathy

