Characterization of calmodulin in the clam Anodonta woodiana: differential expressions in response to environmental Ca2+ and Cd2+
-
Xichao Xia
, Guina Liang
Abstract
Aims
To explore effect of Ca2+ and Cd2+ on the calmodulin (CaM), one complete cDNA sequence (AwCaM1) was cloned and characterized from the freshwater mussel Anodonta woodiana and its expressions were analyzed.
Materials and methods
The AwCaM1 was cloned from the A. woodiana using the rapid amplification of cDNA ends methods and its expression was determined by real-time PCR.
Results
In the hepatopancreas, AwCaM1 expression was up-regulated with a time and dose dependent pattern in the Ca2+ treated groups (0.01, 0.02, 0.04 and 0.08 mg/L) during experiment observed, and increased more than 56.15% (p<0.05) compared with that of control group. AwCaM1 mRNA level increased more 65.04% (p<0.05) in the Cd2+ treated groups (8 and 16 mg/L). In the gill, AwCaM1 expression increased more than 79.41% (p<0.05) compared with that of control group in all the Ca2+ treated groups, and more than 88.23% (p<0.05) in all the Cd2+ treated groups.
Conclusion
These results indicated that up-regulations of AwCaM1 expression in bivalve A. woodiana are associated with Ca2+ absorb and environmental adaption derived from Ca2+ and Cd2+ treatment.
Özet
Amaçlar
Ca+2 ve Cd+2 ‘nin kalmadulin (CaM) üzerine etkisini araştırmak için tatlı su midyesi Anodonta woodiana‘dan tam bir cDNA dizisi (AwCaM1) klonlandı, karakterize edildi ve gen ifadeleri analiz edildi.
Materyal ve Metot
AwCaM1, A. woodiana’dan cDNA uçlarının hızlı amplifikasyonu yöntemi kullanılarak klonlandı ve gen ifadesi gerçek zamanlı PCR ile belirlendi.
Bulgular
Hepatopankreasta AwCaM1 ekspresyonu, zamana ve doza bağımlı olarak Ca+2 ile muamele edilen gruplarda (0.01, 0.02, 0.04 ve 0.08 mg/L) upregüle edildi ve kontrol grubuna kıyasla %56.15’den (p<0.05) fazla arttı. AwCaM1 mRNA seviyesi, Cd+2 ile muamele edilen gruplarda (8 ve 16 mg/L) % 65.04’den (p<0.05) fazla arttı. Solungaçta AwCaM1 ekspresyonu, tüm Ca+2 ile muamele edilen gruplarda kontrol grubuna kıyasla %79.41’den (p<0.05) ve tüm Cd+2 ile muamele edilen gruplarda %88.23’den (p<0.05) fazla arttı.
Sonuç
Bu sonuçlar, A. woodiana’da AwCaM1 ekspresyonunun up-regülasyonlarının, Ca+2 ve Cd+2 muamelesinden kaynaklanan Ca+2 emilimi ve çevre adaptasyonu ile ilişkili olduğunu gösterdi.
Introduction
An array of test organisms, including amphipods, polychaetes, mollusks, crustaceans and fish have been recommend for use in sediment toxicity tests [1]. While this array of test organisms covers a diverse group of fauna, many of them may not be indigenous to all geographic locations [2]. As a group of sedimentary organism, the bivalve filter feeds on suspended particulates and phytoplankton [2]. Their habitat is vulnerable to pollution from urban and industrial development. In addition, adults do not migrate, repopulation of over-fished hard-clam beds depends on the transport of larvae from other areas and several years are required for growth, maturation and reproduction [3]. Using water concentrations associated with response of organism as an indicator of sensitivity, bivalves appear more sensitive to environmental pollution than do other invertebrates [4]. Freshwater clams, Anodonta woodiana are widely distributed in the world and functions as a main criteria required for a bio-indicator organism. Earlier studies have revealed the ability of A. woodiana to accumulate trace elements and pesticides, as well as its potential to detect genotoxicity [5]. In addition, these mussels have important ecosystem functions such as particle filtration and processing, nutrient release and sediment mixing [6]. Unfortunate, metal pollution has resulted in a tremendous threat on their population that can profoundly affect food chain and aquatic ecosystem. Cadmium (Cd), a non-essential metal element required for many of living organisms, can easily enter the environment as a pollutant from many anthropogenic activities [7]. Now, Cd is one of seven most common released heavy metals for the environment (Cd, Cr, Cu, Hg, Ni, Pb and Zn) and its existence causes a serious threat on many invertebrates in aquatic environment, such as crustacean, gastropod mollusks and bivalve [8].
Great evidences have demonstrated intracellular Ca2+ mediates a large number of cellular responses with the high affinity and specificity required for a regulatory second messenger [9]. Calmodulin (CaM), a Ca2+-binding protein, is a ubiquitous, highly conserved, eukaryotic protein that binds and regulates a number of diverse target proteins involved in programmed cell death, autophagy, muscle contraction, inflammation and the immune response [9]. When bound to Ca2+, CaM is able to further bind to CaM-binding proteins (CaMBPs) and directly regulate activities of target proteins [9]. Through the action of these CaMBPs, CaM is involved into a great variety of cellular processes, such as protein translation, protein phosphorylation, cell cycle and cyclic nucleotide metabolism [10]. Notably, several lines of in vitro evidence indicate Cd2+, which has a similar ionic radius to Ca2+, can also bind CaM influencing these downstream effector proteins [11]. A recent in vitro study using osteoblasts derived from rat fetal, demonstrates that Cd2+ treatment significantly increases intracellular Ca2+, leads to CaM activation and ultimately apoptotic death of cells [12]. Other studies specifically implicate the CAMKII pathway as being activated by Cd2+ exposure resulting in apoptosis in cultured mesangial and neuronal cells [13].
Studies on Cd2+-induced toxicological effects have been underdone in the vertebrate, but the mechanisms underlying of observed responses are needed to further elucidate in the invertebrate. Take into consideration of great interest to investigate the effect of metals on A. woodiana, in the current study, one complete cDNA sequence and two premature termination codon mutations of CaM have been cloned and respectively named AwCaM1, AwCaM2 and AwCaM3. In addition, spatio-temporal expressions of AwCaM1 derived from Ca2+ and Cd2+ exposure were determined by quantitative real-time PCR. The present study should be helpful to elucidate the regulatory network of AwCaM1 derived from Ca2+ and Cd2+ treatment. Meanwhile, it also contribute to government paid attention to heavy metals diffusion and take measures to mitigate extensive negative impacts on freshwater organisms and conserve aquatic organism biodiversity.
Materials and methods
Ethics statement
All handling methods of clams were conducted in accordance with the guidelines on the care and use of animals for scientific purposes set up by the Institutional Animal Care and Use Committee of Pingdingshan Medicine College, Pingdignshan, China.
Materials
Approximately 1-year-old of clams A. woodiana (shell length, 6.5±0.5 cm) were obtained from the Baihe River of Nanyang, Henan Province, China. Prior to experiment, animals were maintained in a recirculation system containing filtered freshwater at 24°C for 2 weeks in laboratory. The experiment was conducted in rectangular plastic boxes (40 cm×25 cm; 10 cm height). Clams were cultured in 10 L artificial pond water containing 48 mg NaHCO3, 33 mg CaCl2·2H2O, 60 mg MgSO4·7H2O and 0.5 mg KCl per 1 L deionized water, with a pH of 7.0 [14]. In order to determine the tissue distribution of AwCaM1, five clams derived from same tank were dissected, and several of tissues including foot, gill, hepatopancreas, adductor muscle, heart, hemocytes and mantle were sampled prior to the treatment.
In a Ca and Cd concentration gradient experiment, 330 clams were randomly divided into 11 groups. Each group of 30 specimens was placed into three replicate tanks (10 clams per tank). One group of 30 was used as the control, and the other 10 groups were respectively treated with different concentrations of Ca (as CaCl2, 0.01, 0.02, 0.04, 0.08 and 0.16 mg/L) and Cd (as CdCl2, 1, 2, 4, 8 and 16 mg/L) [15, 16]. In the Ca2+ and Cd2+ incubation treatments, water was replaced with a fresh solution every day in order to assure both the water quality and a constant Ca2+ and Cd2+ content in the medium. Five individuals of each treatment group were randomly sampled at 0, 6, 12, 24, 48 and 72 h, and the hepatopancreas, gill and mantle were collected and stored at −80°C for quantitative real-time reverse transcriptase PCR (qRT-PCR).
Siphoning behavior
The siphoning rate was measured using a previously described method [17] with slight modifications and was based on depletion of neutral red dye particles from the water due to filtration by the clams. Immediately after Ca2+ and Cd2+ exposure, five clams A. woodiana from each Ca2+ and Cd2+ concentration group and control group were placed in 200 mL beakers that contained 100 mL of a neutral red solution and were allowed to siphon for 2 h. Just prior to placing the clams A. woodiana in the solution and just after the 2 h siphoning period, 1 mL aliquots of the water were removed from each beaker, and the neutral red concentration was determined by measuring the optical density at 530 nm using a spectrophotometer. Standard solutions of neutral red were used to generate a standard curve from which the dye concentrations in each test sample were calculated using the equation: where M is the volume of the test solution, n is the number of clams used, t is the time in hours, C0 is the initial concentration of the dye, Ct is the concentration of the dye at time t, and m is the filtration rate
Total RNA isolation and reverse transcription
Total RNA was extracted using TRIzol (Invitrogen Life Technologies, USA) according to the manufacturer’s protocol. Quality of RNA was monitored by 1.2% agarose gel electrophoresis and those with complete rRNA bands were selected to produce cDNA. First-strand cDNA was synthesized using M-MLV First-Strand cDNA synthesis Kit (Takara, China) according to the manufacturer’s instructions and used as the template for PCR reaction.
Cloning of AwCaM cDNA
CaM fragment was amplified using two degenerate primers CaM1 and CaM2 (Table 1) designed according to conserved domains of CaMs of other species including bivalve, gastropod, insect, crustacean and vertebrate. The PCR products were subcloned into the pMDT-19 (Takara, China), sequenced from both directions (Invitrogen Life Technologies, China) and identified CaM partial cDNA sequence. Highly stringent primers (Table 1) designed from the partial cDNA sequences were used to characterize the 5′ and 3′ region of the AwCaMs cDNA by rapid amplification of cDNA ends (RACE) approaches (Takara, China) according to the manufacturer’s protocol. 5′ RACE Outer primer and AwCaM5-1 (Table 1) were used for the first-round PCR of AwCaM 5′ RACE, 3′ RACE Outer primer and AwCaM3-1 (Table 1) for 3′ RACE. Subsequently, 5′ nested PCR was performed by the 5′ first-round PCR product used as template, 5′ RACE Inner primer included in the kit and AwCaM5-2, 3′ nested PCR using 3′ first-round PCR products, 3′ Race Inner primer and AwCaM3-2 (Table 1). The 5′ RACE and 3′ RACE nest PCR products were cloned and five clones were sequenced using the method described above.
Description of the primes used in this study.
| Primer | Sequence (5′–3′) |
|---|---|
| CaM1 | CCATCNCTACNAAGNAANTGGG |
| CaM2 | CTGCTNCACT NATANAGCNA TTTC |
| 5′ RACE Inner primer | CATGGCTACATGCTGACAGCCTA |
| 5′ RACE Outer primer | CGCGGATCCACAGCCTACTGATGATCAGTCGATG |
| AwCaM5-1 | GTTCGGCCTCTGTTGGATTTTGTCCC |
| AwCaM5-2 | GTTCCATCCCCGTCCTTGTC |
| 3′ RACE Outer primer | TACCGTCGTTCCACTAGTGATTT |
| 3′ RACE Inner primer | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| AwCaM3-1 | CTCGGGGAAAAGCTCACAGACGAGG |
| AwCaM3-2 | AGCAGATATTGACGGAGATGG |
| AwCaM-F | CAACAGAGGCCGAACTTCAG |
| AwCaM-R | CCTCGTCTGT GAGCTTTTCC C |
| β-F | CATCCCTTGCTCCTCCAACTATG |
| β-R | CTGGAAGGTAGAGAGAGAAGCCAAG |
CaM1 and CaM2 are degenerate primers and used to isolate partial cDNA of CaM. 5′ RACE Outer primer, 5′ RACE Inner primer, AwCaM5-1 and AwCaM5-2 are used to characterize the 5′ RACE of the AwCaM in the nest PCR, 3′ RACE Outer primer, 3′ RACE Inner primer, AwCaM3-1 and AwCaM3-1 for 3′ RACE. AwCaM-F and AwCaM-R as well as β-F and β-R are selected to isolate AwCaM1 and β-actin in real-time PCR, respectively.
Sequence and phylogenetic analysis
AwCaMs sequence were analyzed and compared using the BLAST program with a GenBank database search (www. ncbi.nlm.nih.gov/blast). The signal peptide was predicted by signal program (http://www.cbs.dtu.dk/services/SignalP). Prediction of protein domain was carried out with the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/). Multiple sequence alignments of AwCaMs gene were performed using the DANMEN analysis program. Prediction of three-dimensional (3D) structure was fulfilled by Swiss-model (http://swissmodel.expasy.org/). Phylogenetic trees constructed from the alignment were generated by the Neighbor-joining method using MEGA5.0 software. Reliability of trees obtained was assessed by bootstrapping, using 1000 bootstrap replications.
Quantification of AwCaM1 expression by real-time PCR
To determine the mRNA levels of AwCaM1 derived from hepatopancreas and other tissues, real-time quantitative PCR was underdone following the manufacture instruction of SYBR Premix Ex TaqTM (TaKaRa, China). Firstly, AwCaM1 primers as well as β-actin primers (Table 1) were designed based on isolated sequences of A. woodiana, respectively, used to isolated target genes in common PCR instrument, and only one band was detected in the PCR production by agarose gel electrophoresis. PCR products were sequenced and identified as the partial sequence of target genes. Subsequently, real-time PCR was fulfilled using an ABI 7500 Real-Time Detection System (Applied Biosystems, USA). Based on constructed standard curve, expression levels of AwCaM1 were calculated by 2-∆∆CT. All data were given in terms of relative mRNA expression as means±SE.
Statistical analyses
All of the experimental data analyses were subjected to one-way analysis of variance (ANOVA) and p<0.05 was considered statistically significant.
Results
Characterization of AwCaM1 cDNA
The complete cDNA sequence of AwCaM1 was obtained and deposited in GenBank under accession number KY996397, and comprised of 692 bp, including a 5′ untranslated region (UTR) of 21 nucleotides, a 516 bp open reading frame (ORF) which was encoded 172 amino acids, and a long 3′-UTR of 155 nucleotides containing a stop codon (TAG), a putative polyadenylation consensus signal (AATAAA), and a poly (A) tail (Figure 1). The calculated molecular mass of deduced mature AwCaM1 was 19.22 kDa and had a theoretical isoelectric point of 4.07. AwCaM1 contained four putative Ca2+-binding EF-hand motifs which were respectively located at 34–63 aa, 71–99 aa, 108–136 aa and 144–172 aa (Figure 1). Comparing the variance of amino acid sequences, the conserved amino acid residues were highly fund in AwCaM1 and CaM of others. Two crucial phosphorylation sites of the human CaM (Thr79 and Ser81) were respectively identified in AwCaM1 (102 aa and 104 aa) (Figure 2). Notably, alignment result showed only three residues of Arg75 and Ile86 and Tyr100 in human were respectively substituted by Lys, Leu and Phe in AwCaM1 (Figure 2).
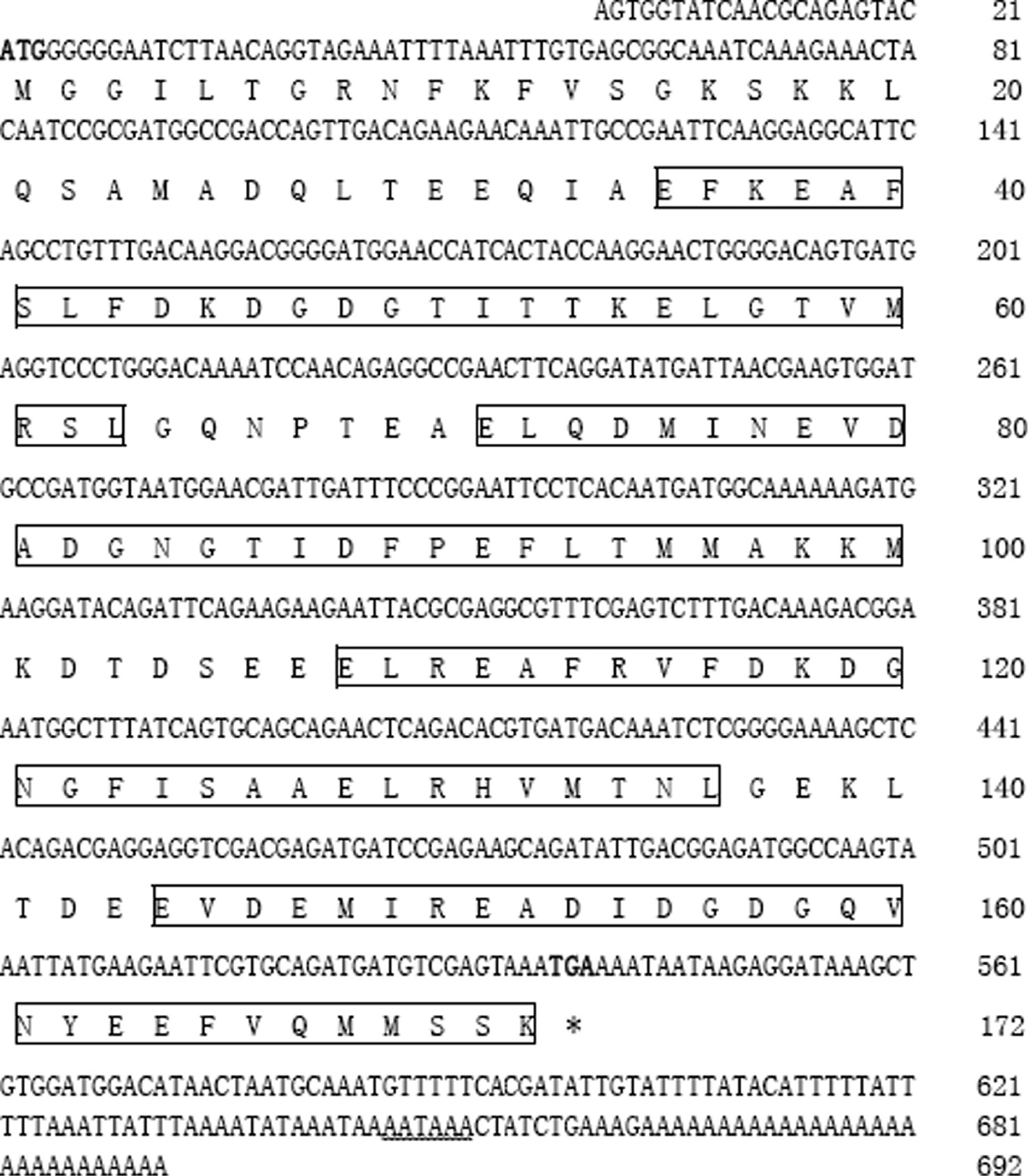
Nucleotide and deduced amino acid sequences of AwCaM1.
The start and stop codons are indicated with bold. Putative polyadenylation signal “AATAAA” is showed with wavy line. The four Ca2+-binding domains are marked with box.
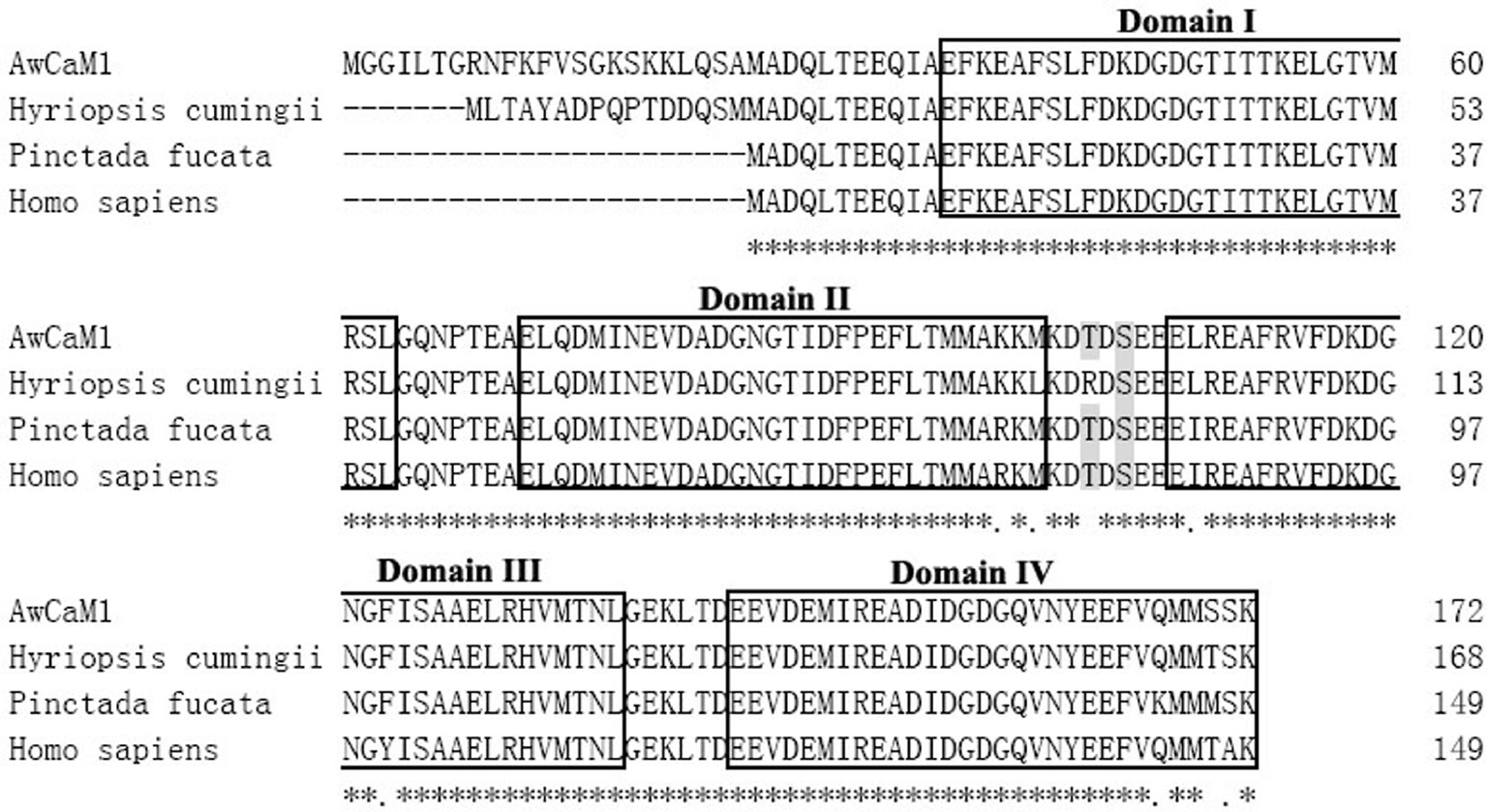
Multiple alignment of AwCaM1 with other CaMs.
Two crucial phosphorylation sites of Thr and Ser are indicated with shadow. The four Ca2+-binding domains are marked with box.
Furthermore, predicted secondary structure of AwCaM1 protein contained seven alpha-helices and eight β-sheets (Figure 3A) that was very similar to CaM secondary structure of other species, especially to that of human. 3D structure of AwCaM1 showed a high-degree similarity with CaMs (Figure 3B).
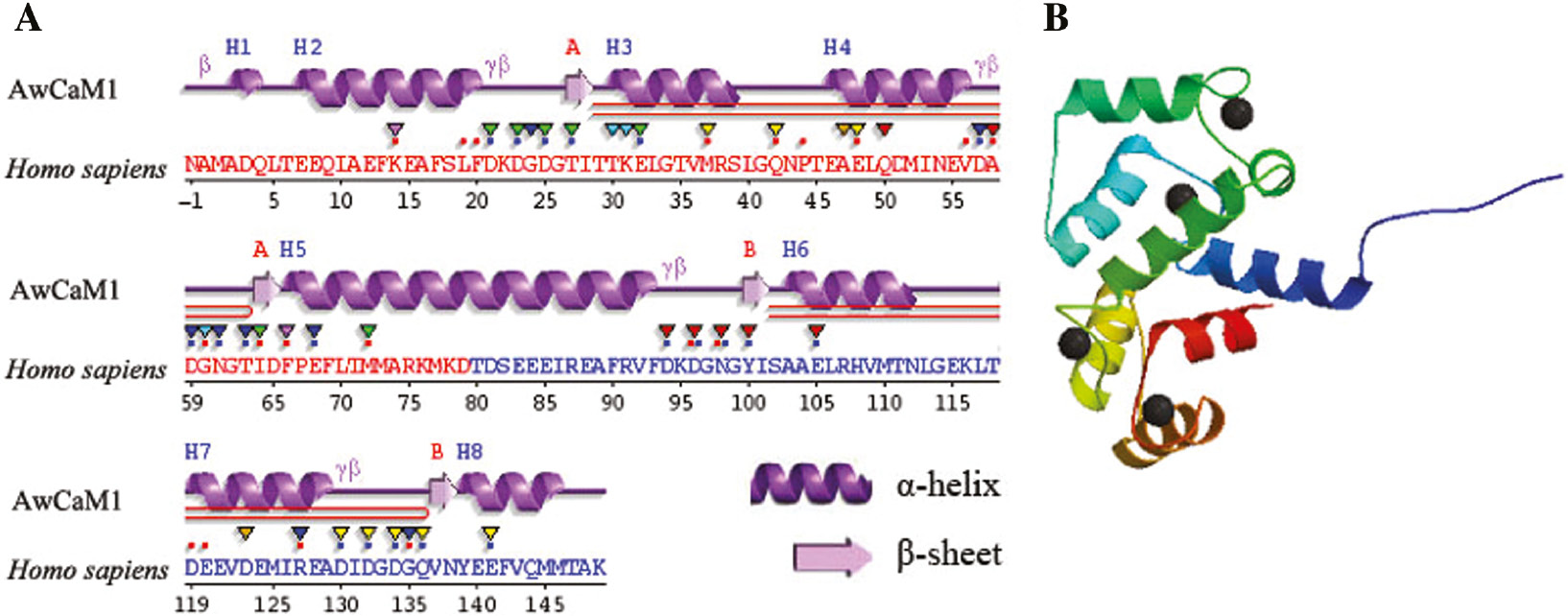
Predicted secondary and 3D structures of AwCaM1 deduced amino acids.
(A) The secondary structure of AwCaM1. (B) The 3D structure of AwCaM1.
Evolutionary relationship of AwCaM1
BLAST analysis revealed that amino acid sequence of AwCaM1 was close with other CaMs. The overall deduced amino acid sequence of AwCaM1 exhibited 96% to freshwater bivalve Pinctada fucata, 94% to insect Daphnia magna, 96% to Drosophila melanogaster and 95% with Homo sapiens. A molecular phylogenetic tree was generated to further analyze the evolutionary relationships between AwCaM1 and other CaMs sequences (Figure 4). In evidenced sequences of mollusk, AwCaM1 shared the higher relationship of evolution with clam of freshwater. Interestingly, the freshwater calms and seawater ones were not grouped in one cluster although they were belonged to same class (Figure 4). In addition, species were not orderly arranged according to evolutional relationship, such as spider Cupiennius salei, holothuroidea Apostichopus japonicas, fish Branchiostoma belcheri tsingtauense located in one cluster as well as chordata Ciona intestinalis, camel Camelus dromedaries, Homo sapiens, bird Cariama cristata distributed in one clade (Figure 4). Notably, few lower bootstrap values were observed between two classes (Figure 4).

Phylogenetic relation of AwCaM1 from other organisms.
AwCaM1 is marked with underline. Crassostrea gigas (accession number XP_011436578.1), Ciona intestinalis (accession number NP_001027633.1), Cupiennius salei (accession number CFW94154.1), Drosophila melanogaster (accession number NP_523710.1), Papilio xuthus (accession number KPI95926.1), Caenorhabditis elegans (accession number NP_503386.1), Procambarus clarkii (accession number ACI15835.1), Camelus dromedarius (accession number XP_010996006.1), Cariama cristata (accession number XP_009702107.1), Pinctada fucata (accession number JAS03530.1), Litopenaeus vannamei (accession number AEK21539.1), Hyriopsis cumingii (accession number ADT61781.1), Apostichopus japonicus (accession number AAY41437.1), Bombyx mori (accession number XP_012546645.1), Homo sapiens (accession number NP_001734.1), Branchiostoma belcheri tsingtauense (accession number ABM53481.1).
Analysis of two premature termination codon mutations of AwCaM1
Two premature termination codon mutations of CaM from A. woodiana were isolated and named as AwCaM2 (GenBank accession number KY996398) and AwCaM3 (GenBank accession number KY996399), their full cDNA lengths were 610 bp and 546 bp, respectively. The AwCaM2 and AwCaM3 share similar sequence composition with AwCaM1 from 1st to 535th bp (Figure 5). In contrasted with cDNA sequence of AwCaM1, AATAA signal sequence was missed in 3′ UTR of AwCaM2, stop-code and AATAA signal sequences lacked in the AwCaM3 (Figure 5). Therefore, AwCaM2 and AwCaM3 were the individuals of premature termination codon mutations of CaM transcription.

Alignment of nucleotide sequences of AwCaM1, AwCaM2 and AwCaM3.
Stop codons (TGA) are indicated with green box. Putative polyadenylation signal “AATAAA” is showed with red underline.
Tissue distribution of AwCaM1
Quantitative real-time PCR was employed to investigate the distribution of AwCaM1 in different tissues. The constitutive expression levels of AwCaM1 were examined in different tissues including foot, mantle, adductor muscle, heart, hepatopancreas, hemocytes and gill, and showed different expression profiles (Figure 6). Higher expressions of AwCaM1 were observed in mantle and gill, a moderate level in hepatopancreas, foot and heart, but a lower level in adductor muscle and hemocytes (Figure 6).

Real-time PCR analysis of AwCaM1 transcript from different tissues.
n=3 Replicates.
Ca2+ and Cd2+ effect on the siphoning behavior
No statistical difference of the siphoning behavior was observed between the control group and lower concentration Ca2+ treated groups (0.01, 0.02, 0.04 and 0.08 mg/L) (Figure 7). However, the filtration rate at 0.16 mg/L Ca2+ (24.84±3.04 mL/animal/h) was significantly decreased compared with that of the control group (38.38±3.12 mL/animal/h) (p=0.0039) (Figure 7). In addition, administration different concentrations of Cd2+ could result in a decrease of filtration rate (Figure 7). Compared with that of control group, filtration rate of individual clam reduced more than 31.37% (p=0.0106), 56.04% (p=0.0006), 64.35% (p=0.0003) and 87.38% (p=0.0001) in the 2, 4, 8 and 16 mg/L of Cd2+ treated group, respectively.
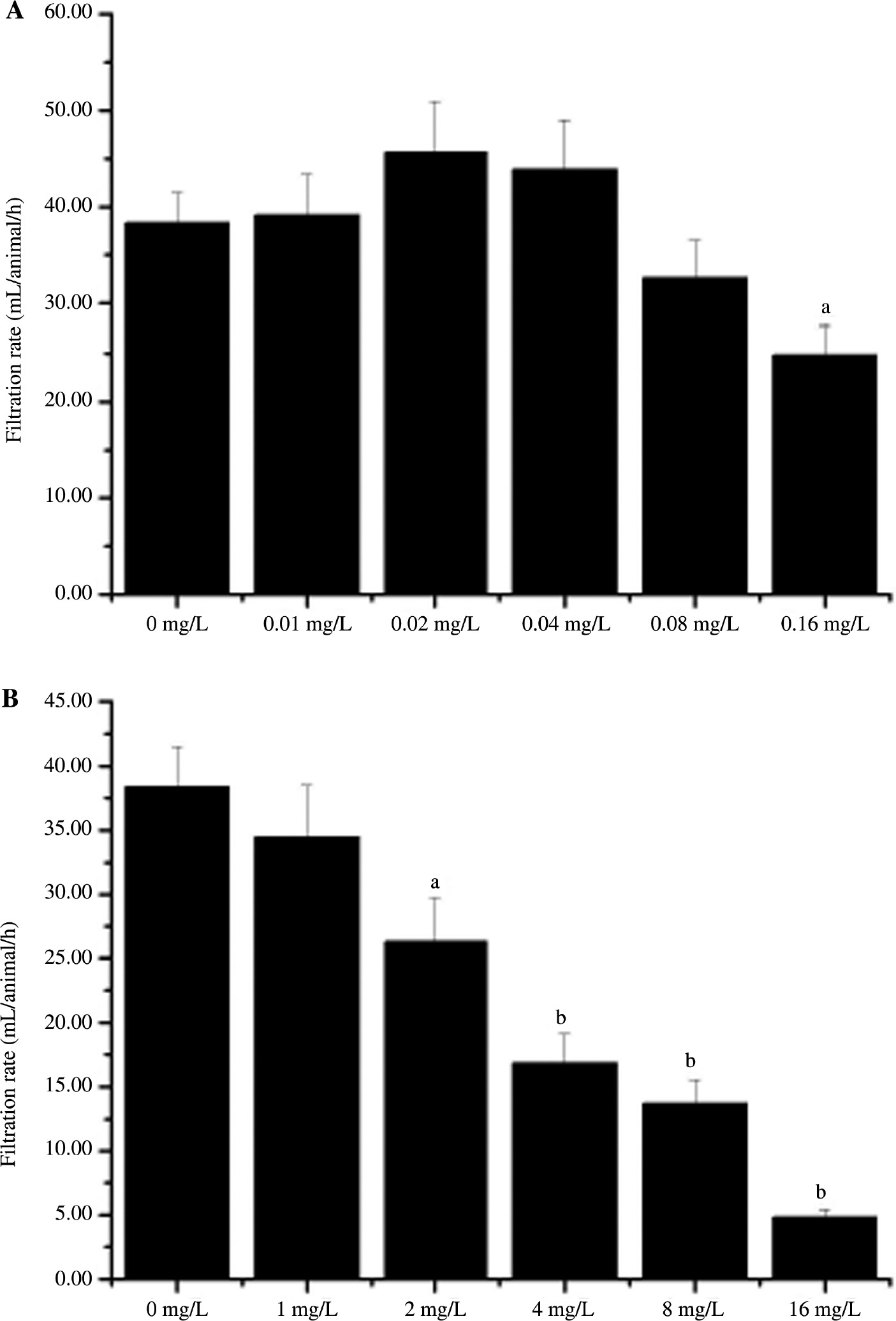
Filtration rate (mL/animal/h) of Anodonta woodiana exposed to Ca2+ and Cd2+ treated water.
Average (n=5) filtration rate (mL/animal/h) of clams in Ca2+ and Cd2+ treated water containing a 1 mg/L neutral red solution (0–50 mg/L) following a 72-h exposure period to the concentrations indicated. The experiments were performed in triplicate and repeated three times with similar results. The bars display the mean±SE. A statistically significant difference between treatment group and control (p<0.05) is indicated by differences in the letters above the bars. ap<0.05, bp<0.01 vs. control group at the treated group.
Temporal expressions of AwCaM1 in hepatopancreas exposured to Ca2+ and Cd2+
Temporal expressions of AwCaM1 mRNA in hepatopancreas were significantly affected by Ca2+ and Cd2+ challenge using RT-qPCR with β-actin as internal control (Figure 8). In the Ca2+ treated groups (0.01, 0.02, 0.04 and 0.08 mg/L), profiles of AwCaM1 expression showed a time and dose dependent pattern with respect of that of control group during experiment observed (Figure 8A). Meanwhile, in the Ca2+ treated groups (0.08 and 0.16 mg/L), AwCaM1 expression increased more than 56.15% (p=0.0003) from 6 h to 72 h (Figure 8A). In the Cd2+ treated groups (1, 2 and 4 mg/L), up-regulation of AwCaM1 expression showed a timed and dose dependent patter compared with that of control group (Figure 8B). In the Cd2+ treated groups (8 and 16 mg/L), AwCaM1 mRNA levels increased more 65.04% (p=0.0001) from 6 h to 72 h (Figure 8B).

Temporal expressions of AwCaM1 in hepatopancreas after Ca2+ and Cd2+ challenge as measured by quantitative real-time PCR.
(A) Effect of Ca2+ on AwCaM1 expression in hepatopancreas. (B) Effect of Cd2+ on AwCaM1 expression in hepatopancreas. Bars represented means±SE; n=3/each group/each time point. ap<0.05, bp<0.01 vs. control group at the same time.
Temporal expressions of AwCaM1 in gill exposured to Ca2+ and Cd2+
In gill, treatment of different concentrations of Ca2+ caused a significant induction of AwCaM1 expression. Up-regulation of AwCaM1 expressions characterized with a timed and dose dependent patter was observed in the Ca2+ treated groups (0.01, 0.02, 0.04 and 0.08 mg/L) from 6 h to 72 h (Figure 9A). Among of all the Ca2+ treated groups, AwCaM1 expression increased more than 79.41% (p=0.0001) compared with that of control group (Figure 9A). Like mode of Ca2+ treatment, administration of Cd2+ could result in a significant up-regulation of AwCaM1 expression. In all the Cd2+ treated groups, AwCaM1 expression increased more than 88.23% (p=0.0001) in contrasted with that of control group (Figure 9B).

Temporal expressions of AwCaM1 in gill after Ca2+ and Cd2+ challenge as measured by quantitative real-time PCR.
(A) Effect of Ca2+ on AwCaM1 expression in gill. (B) Effect of Cd2+ on AwCaM1 expression in gill. Bars represented means±SE; n=3/each group/each time point. ap<0.05, bp<0.01 vs. control group at the same time.
Temporal expressions of AwCaM1 in mantle exposured to Ca2+ and Cd2+
In the mantle, up-regulation of AwCaM1 expression with a time and dose dependent pattern was observed in the Ca2+ treated groups (0.01, 0.02, 0.04 and 0.08 mg/L) during experiment observed (Figure 10A). With Ca2+ treatment at a concentration of 0.16 mg/L, AwCaM1 expression increased more than 1.69 times (p=0.0001) from 6 h to 72 h compared with that of control group (Figure 10A). In the Cd2+ treated groups (1, 2 and 4 mg/L), up-regulation of AwCaM1 expression showed a timed and dose dependent patter. In the Cd2+ treated groups (8 and 16 mg/L), AwCaM1 mRNA levels increased more 1.65 times (p=0.0001) from 6 h to 72 h in contrasted with that of control group (Figure 10B).

Temporal expressions of AwCaM1 in mantle after Ca2+ and Cd2+ challenge as measured by quantitative real-time PCR.
(A) Effect of Ca2+ on AwCaM1 expression in mantle. (B) Effect of Cd2+ on AwCaM1 expression in mantle. Bars represented means±SE; n=3/each group/each time point. ap<0.05, b p<0.01 vs. control group at the same time.
Discussion
CaM is a pivotal calcium metabolism regulator and involved into the shell formation process of the pearl oyster. In current study, one complete cDNA sequence of CaM gene was isolated from freshwater bivalve A. woodiana and named AwCaM1, and its expression was also examined in different tissues and different times.
The deduced primary amino acid sequence of AwCaM1 showed a high identity with the published CaMs of a range of invertebrate and vertebrate species. With respect of difference of amino acid composition, only three amino acid residues were observed between AwCaM1 and human CaM. Meanwhile, the CaM genes from different vertebrate species encode the same CaM molecule with high identical amino acid sequences. In current study, AwCaM1 was also identified with that from crustacean D. magna, and insect D. melanogaster, which indicates that the CaM proteins are highly conserved during invertebrate evolution. Notably, residue substitution, an interesting phenomenon, was detected in the present study. Compared to the marine pearl oyster P. fucata CaM, there is a single-amino acid substitution at position 147 in which Met is occupied by the Thr in freshwater bivalve Hyriopsis cumingii and Ser in A. woodiana, respectively. The Met at this position in P. fucata is considered to be a characteristic residue of the oyster [18], but it is not found in these freshwater clams. What reasons of substitution between oyster and freshwater clams are needed be further elucidate. In addition, compared with the deduced protein sequences, an important phosphorylation site Tyr100 in the Ca2+-binding ligand position of the third EF-hand motif of human Cam was substituted by Phe100 in AwCaM1. It has demonstrated that phosphorylation of Tyr100 in human works as important role of increase binding affinity for its target proteins [19]. The substitution of phosphorylation sites of 100 aa is likely associated with the regulation of AwCaM1 at the posttranscriptional level. On another hand, as a key binding site of EF-hand motif, event of substitution should result in a conformational change of this protein that further effect its affinity for most of its target proteins and its ability to activate target proteins. To shed light of these phenomena is a very interesting work in the future.
CaM, a multifunctional Ca2+ binding protein, is involved in the regulation of numerous important physiological functions, including neural activity, gene expression, enzyme regulation and muscle contraction. CaM is highly conserved across different species, and comprised of four EF hands that generally form two structurally similar domains connected by a flexible central linker. Although CaMs of these species are highly conserved and share similar structure, phylogenetic result showed that few lower bootstrap values were observed in different clusters in these CaMs located. An obvious distance of phylogenetic relationship was also detected between freshwater calms and seawater ones in spite they all belong to one class. Meanwhile, different species derived from vertebrate and invertebrate were grouped into similar cluster. Based on mentioned above, it is suggested CaM is not a candidate for reflecting the phylogenetic relationship, but an important player for sustaining normal physiological function of animals.
In all the tissues, strong expression of AwCaM1 was observed in mantle, suggested that is associated with the shell formation. The original hypotheses indicate that CaM expression should be greater in epithelial tissues (such as the gill and mantle) and lesser in non-epithelial tissues (such as muscle) [20]. In mollusks, the mantle, especially the outer epithelium of the mantle, plays a key role in shell formation [21]. In current study, the AwCaM1 mRNA expression level was obvious higher in the mantle compared to the adductor muscle. Similar results are also reported from other animals. In addition, the higher expression level of AwCaM1 mRNA was observed in the gill that supports previous conclusion in which gill is considered as a key organ of calcium uptake and accumulation in the bivalve. It has demonstrated that the gill is a primary organ for calcium uptake from the water in bivalves. Particularly in the scallop, the gill is thought to play a regulatory role in the membrane Ca2+-ATPase system or to act as a “calcium sink” [18].
In the current study, AwCaM2 and AwCaM3, two premature termination codon mutations of CaMs, were also isolated from A. woodiana, suggesting there is a complex mechanism involved into controlling AwCaM1 transcription. As to phenomena, studies of loss-of-function mutations pave an avenue to elucidate mechanism, it has gain great attention. Loss-of-function mutations are generally believed to be deleterious and have been discussed in the context of human medicine [22]. Now, genome wide surveys in human and fly populations reveal an unexpected prevalence of loss-of-function mutations with hundreds or thousands of genes harboring deletions and/or premature termination codon mutations [23]. These genes are narrowly transcribed and encode high numbers of paralogs. But, gene expression in eukaryotes is a compartmentalized process consisting of several different, yet connected steps. In the process, importance of gene expression for living cells and organisms is exemplified by the existence of diverse molecular mechanisms that detect errors and thereby ensure the accuracy of gene expression. A well-known quality control process, referred to as nonsense-mediated mRNA decay or alternatively mRNA surveillance, limits the expression of mRNAs with premature termination codons and other aberrant termination events. So, in order to tolerate existence of these genes, a sophisticated mechanism should be involved in the regulated process of transcription [22, 23].
Filtration rates of Cd2+ treated groups were significant reduced compared with that of control group, suggesting that treatment of Cd2+ could result in an obvious injury on the health clams. Combination of siphoning behavior and valve movement has been utilized as indicators for continuous biomonitoring of water supplies and effluents [24]. Bivalve siphons play important roles in nutritional physiology, defense, and reproduction and are therefore a signal of general healthy or stress. Previous studies have demonstrated that bivalve siphoning can be influenced by metals or chlorpyrifos [17, 25]. Valves of the clams are always closed during exposure to 3.13 mg/L chlorpyrifos treatments [26]. In addition, a reduction of siphoning activity as a response to chemical stress has also been observed that is associated with ammonia accumulation in the tissue, a reduction in oxygen exchange, and reduced feeding which could all have significant implications for survival, growth and reproduction of bivalves [25, 27]. Herein, we found that Cd2+ exposure decreased the filtration rates of clams, although no significant valve closure was observed. It is therefore reasonable to conclude that the decreased siphoning caused by Cd2+ exposure reflects an overall negative health impact and an indicator of chemical stress.
In the present study, treatment of different Ca2+ concentrations could result in a significant up-regulation of AwCaM1 expression in the hepatopancreas, gill and mantle, suggested that stimulating AwCaM1 transcription might help A. woodiana to maintain cytosolic Ca2+ homeostasis and gain compensatory Ca2+ absorption as well as adapt to environmental stress. CaM plays a pivotal role in cellular Ca2+ homeostasis by activating Ca2+-ATPase, which is the major transporter in Ca2+ fluxes [15, 28]. Up-regulation of AwCaM1 is contributed to increase Ca2+ absorption of cells. On another hand, in these tissues, expression of AwCaM1 was down-regulated at 72 h compared with that of 48 h in the Ca2+ treated group at a concentration of 0.16 mg/L. Significant up-regulation of AwCaM1 expression is also associated with coping environmental stress derived from Ca2+ treatment, especially in the higher concentration. Expression of CaMs of animals could be induced by environmental stressors, such as hypo-osmotic stress and pathogenic organism [21, 29]. Evidence has indicated that CaM is considered as one of the most important molecular biomarkers in teleost exposed to the chronic stressors [30]. Previous studies demonstrated that the cold adaptation in Dissostichus mawsoni is highly related to CaM expression [31]. Given that CaM is an important subunit for several ion channels, the significant up-regulation of AwCaM1 expression likely function as dual role in the different concentrations of Ca2+ treatment.
In the hepatopancreas, gill and mantle, up-regulations of AwCaM1 with a time and dose dependent pattern were occurred in the Ca2+ treated groups at concentrations from 0.01 to 0.08 mg/L. In addition, filtration rates of Ca2+ treated groups showed a gradually up-regulation trend with respect of increase of treated concentrations between 0.01 and 0.04 mg/L. Combination of characterizations of AwCaM1 expression and filtration rate, we postulated, under 0.04 mg/L, administration of Ca2+ is contribute to increase absorption of Ca2+ and further maintain cytosolic Ca2+ homeostasis as well as boost shell formation of calms.
In the current study, treatment of Cd2+ at different concentrations could result in a significant up-regulation of AwCaM1 expression in the hepatopancreas, gills and mantile, suggested stimulation of AwCaM1 transcription is associated with Cd2+ toxicity. Due to their filtration activity, bivalves are exposed to numerous heavy metals and thus are subject to a considerable amount of oxidative damage [32]. It has found Cd2+ can induced oxidative DNA lesions in bivalves of freshwater and sea water. In the Mytilus edulis, low concentrations of Cd2+ enhance the genotoxicity of H2O2 and Cd2+ inhibits the DNA repair of 8-oxodG [33]. Further study shows that Cd2+ induced an increase of the DNA strand break levels and a low level of 8-hydroxy guanine DNA glycosidase-sensitive sites [34]. Undoubtedly, existence of Cd2+ can cause tremendous threat on cell survival. Notably, what is mechanism of Cd2+ entering into cells? Studies have indicated treatment of Cd2+ can cause break of Ca2+ homeostasis, and result in apoptosis of a variety of cells [35]. In the process, some Ca2+ related proteins are significantly up-regulated after Cd2+ exposure, such as neuronal calcium sensor, sarcoplasmic calcium-binding protein and CaM [35]. In addition, it was assumed that Ca2+ allow to be displaced by Cd2+ in some Ca2+-binding proteins with respect of the biophysicochemical similarities of Cd2+ and Ca2+ that disrupt Ca-mediated signaling pathways. Notably, research indicates Cd2+ exposure can lead to a transient rise in intracellular Ca2+ ions in several cell types, especially in osteoblasts [12]. Osteoblasts can uptake Cd2+ ions via membrane-bound transporters, such as transient receptor potential channels [36]. Consequently, it is possible that a combination of increased intracellular Cd2+ and Ca2+ ions can bind CaM leading to activation or inhibition of Ca2+/calmodulin dependent kinase (CAMK) pathways. Other studies specifically implicate the CAMKII pathway as being activated by Cd2+ exposure that result in apoptosis in cultured mesangial and neuronal cells [37]. However, the roles of the other two pathways, CaM-dependent PDE and CAMKK, in Cd2+ toxicity are under-investigated. Taken together, these studies provide evidence in support of the current research. Therefore, up-regulation of AwCaM1 is related with accumulation of Cd2+ that should lead to a risk of cell damage.
Notably, the AwCaM1 expressions of hepatopancreas, gill and mantle showed a dose- and time-dependent manner in lower concentration of Cd2+ treated groups. However, AwCaM1 expression showed a biphasic profile in the 8 and 16 mg/L of Cd2+ treated groups characterized by up-regulation matter in the early stage and down-regulation trend in the latter stage. This pattern of AwCaM1 is associated with persistent accumulation of reactive oxygen species (ROS) derived from Cd2+ injury. With elongation of Cd2+ treated time, ROS should increasingly be produced and accumulated in the cells. Levels of abruptly increased ROS is exceed the eliminating ability of cells that cause redox imbalance possibly via a shift between oxidants and antioxidants in favor of oxidants, and result in chronic inflammation, apoptosis of immune cells, etc. [38]. In the condition, if animals cannot produce new cells to compensate for death ones, number of living cells should be gradually decreased. Following, expressions of AwCaM1 are also decline alone treated time. Thus, a biphasic phenomenon was detected in higher concentration groups of Cd2+.
In conclusion, one complete cDNA sequence and two premature termination codon mutations of CaMs (AwCaM1, AwCaM2 and AwCaM3) were cloned and characterized from the freshwater mussel A. woodiana. With respect of complex process from binding CaM to activation, effect of Ca2+ and Cd2+ on the freshwater bivalve A. woodiana is an sophisticated event. Use of AwCaM1 expression is contributed to provide insight into the important landscape of change at the molecular level derived from Ca2+ and Cd2+ treatment. This work is helpful to elucidate the potential mechanism of Cd2+ toxicity and provide a key regulator of Cd2+ toxicity in freshwater clam A. woodiana. The continuous functional investigation of proteins of network into Ca2+ involved could eventually solve the key targets of Cd2+ toxicity. Meanwhile, government should concern heavy metals diffusion and take measures to mitigate extensive negative impacts on freshwater organisms and conserve aquatic organism biodiversity. Notably, the present work is only the laboratory results. So, great efforts are required to explore the application of A. woodiana in the environmental engineering and environmental monitoring.
Acknowledgment
This research was funded by the National Natural Science Foundation of Henan (No. 18A330004, PXY-BSQD-2018009, 2015GGJS-286, 17A180010) and China Postdoctoral Science Foundation Funded Project (2016M590143).
References
1. Akpheokhai LI, Oribhabor BJ. Nematodes relevance in soil quality management and their significance as biomarkers in aquatic substrates: review. Recent Pat Biotechnol 2016;10:228–34.10.2174/1872208310999160811164838Suche in Google Scholar PubMed
2. Tiling K, Proffitt CE. Effects of Lyngbya majuscula blooms on the seagrass Halodule wrightii and resident invertebrates. Harmful Algae 2017;62:104–12.10.1016/j.hal.2016.11.015Suche in Google Scholar PubMed
3. Le TT, Zimmermann S, Sures B. How does the metallothionein induction in bivalves meet the criteria for biomarkers of metal exposure? Environ Pollut 2016;212:257–68.10.1016/j.envpol.2016.01.070Suche in Google Scholar PubMed
4. Suárez-Ulloa V, Fernández-Tajes J, Manfrin C, Gerdol M, Venier P, Eirín-López JM. Bivalve omics: state of the art and potential applications for the biomonitoring of harmful marine compounds. Mar Drugs 2013;11:4370–89.10.3390/md11114370Suche in Google Scholar PubMed PubMed Central
5. Zhang X, Liu Z, Jeppesen E, Taylor WD. Effects of deposit-feeding tubificid worms and filter-feeding bivalves on benthic-pelagic coupling: implications for the restoration of eutrophic shallow lakes. Water Res 2014;50:135–46.10.1016/j.watres.2013.12.003Suche in Google Scholar PubMed
6. Chen J, Xie P. Seasonal dynamics of the hepatotoxic microcystins in various organs of four freshwater bivalves from the large eutrophic lake Taihu of subtropical China and the risk to human consumption. Environ Toxicol 2005;20:572–84.10.1002/tox.20146Suche in Google Scholar PubMed
7. Isani G, Andreani G, Cocchioni F, Fedeli D, Carpené E, Falcioni G. Cadmium accumulation and biochemical responses in Sparus aurata following sub-lethal Cd exposure. Ecotoxicol Environ Saf 2009;72:224–30.10.1016/j.ecoenv.2008.04.015Suche in Google Scholar PubMed
8. Shi W, Zhao X, Han Y, Che Z, Chai X, Liu G. Ocean acidification increases cadmium accumulation in marine bivalves: a potential threat to seafood safety. Sci Rep 2016;6:20197.10.1038/srep20197Suche in Google Scholar PubMed PubMed Central
9. Means AR, Dedman JR. Calmodulin – an intracellular calcium receptor. Nature 1980;285:73–7.10.1038/285073a0Suche in Google Scholar PubMed
10. Zayzafoon M, Fulzele K, McDonald JM. Calmodulin and calmodulin-dependent kinase IIalpha regulate osteoblast differentiation by controlling c-fos expression. J Biol Chem 2005;280:7049–59.10.1074/jbc.M412680200Suche in Google Scholar PubMed
11. Shirran SL, Barran PE. The use of ESI-MS to probe the binding of divalent cations to calmodulin. J Am Soc Mass Spectrom 2009;20:1159–71.10.1016/j.jasms.2009.02.008Suche in Google Scholar PubMed
12. Liu W, Zhao H, Wang Y, Jiang C, Xia P, Gu J, et al. Calcium-calmodulin signaling elicits mitochondrial dysfunction and the release of cytochrome c during cadmium-induced apoptosis in primary osteoblasts. Toxicol Lett 2014;224:1–6.10.1016/j.toxlet.2013.10.009Suche in Google Scholar PubMed
13. Chen S, Xu Y, Xu B, Guo M, Zhang Z, Liu L, et al. CaMKII is involved in cadmium activation of MAPK and mTOR pathways leading to neuronal cell death. J Neurochem 2011;119:1108–18.10.1111/j.1471-4159.2011.07493.xSuche in Google Scholar PubMed PubMed Central
14. Booth A, Zou E. Impact of molt-disrupting BDE-47 on epidermal ecdysteroid signaling in the blue crab, Callinectes sapidus, in vitro. Aquat Toxicol 2016;177:373–9.10.1016/j.aquatox.2016.06.011Suche in Google Scholar PubMed
15. Liu H, Chen X, Su Y, Kang IJ, Qiu X, Shimasaki Y, et al. Effects of calcium and magnesium ions on acute copper toxicity to Glochidia and early juveniles of the Chinese pond mussel Anodonta woodiana. Bull Environ Contam Toxicol 2016;97:504–9.10.1007/s00128-016-1890-8Suche in Google Scholar PubMed
16. Evariste L, Rioult D, Brousseau P, Geffard A, David E, Auffret M, et al. Differential sensitivity to cadmium of immunomarkers measured in hemocyte subpopulations of zebra mussel Dreissena polymorpha. Ecotoxicol Environ Saf 2017;137:78–85.10.1016/j.ecoenv.2016.11.027Suche in Google Scholar PubMed
17. Cooper NL, Bidwell JR. Cholinesterase inhibition and impacts on behavior of the Asian clam, Corbicula fluminea, after exposure to an organophosphate insecticide. Aquat Toxicol 2006;76:258–67.10.1016/j.aquatox.2005.09.012Suche in Google Scholar PubMed
18. Li S, Xie L, Ma Z, Zhang R. cDNA cloning and characterization of a novel calmodulin-like protein from pearl oyster Pinctada fucata. FEBS J 2005;272:4899–910.10.1111/j.1742-4658.2005.04899.xSuche in Google Scholar PubMed
19. Corti C, Leclerc LE, Quadroni M, Schmid H, Durussel I, Cox J, et al. Tyrosine phosphorylation modulates the interaction of calmodulin with its target proteins. Eur J Biochem 1999;262:790–802.10.1046/j.1432-1327.1999.00441.xSuche in Google Scholar PubMed
20. Tang M, Shi A. Studies of environmental calcium concentration effect on the calcium metabolism of the mantle and pearel sac of the freshwater pearl mussel. J Sichuan Univ 2000;37:741–7.Suche in Google Scholar
21. Ren G, Hu X, Tang J, Wang Y. Characterization of cDNAs for calmodulin and calmodulin-like protein in the freshwater mussel Hyriopsis cumingii: differential expression in response to environmental Ca(2+) and calcium binding of recombinant proteins. Comp Biochem Physiol B Biochem Mol Biol 2013;165:165–71.10.1016/j.cbpb.2013.04.003Suche in Google Scholar PubMed
22. Hug N, Longman D, Cáceres JF. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res 2016;44:1483–95.10.1093/nar/gkw010Suche in Google Scholar PubMed PubMed Central
23. Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 2016;129:461–7.10.1242/jcs.181008Suche in Google Scholar PubMed PubMed Central
24. Chen WY, Jou LJ, Chen SH, Liao CM. A real-time biomonitoring system to detect arsenic toxicity by valve movement in freshwater clam Corbicula fluminea. Ecotoxicology 2012;21:1177–87.10.1007/s10646-012-0872-9Suche in Google Scholar PubMed
25. Moulton CA, Fleming WJ, Purnell CE. Effects of two cholinesterase-inhibiting pesticides on freshwater mussels. Environ Toxicol Chem 1996;15:131–7.10.1002/etc.5620150210Suche in Google Scholar
26. Milam CD, Farris JL. Risk identification associated with iron-dominatedmine discharges and their effect upon freshwater bivalves. Environ Toxicol Chem 1998;17:1611–9.10.1002/etc.5620170824Suche in Google Scholar
27. Chen H, Zha J, Liang X, Li J, Wang Z. Effects of the human antiepileptic drug carbamazepine on the behavior, biomarkers, and heat shock proteins in the Asian clam Corbicula fluminea. Aquat Toxicol 2014;155:1–8.10.1016/j.aquatox.2014.06.001Suche in Google Scholar PubMed
28. Snedden WA, Fromm H. Calmodulin as a versatile calcium signal transducer in plants. New Phytol 2001;151:35–66.10.1046/j.1469-8137.2001.00154.xSuche in Google Scholar PubMed
29. Ji PF, Yao CL, Wang ZY. Two types of calmodulin play different roles in Pacific white shrimp (Litopenaeus vannamei) defenses against Vibrio parahaemolyticus and WSSV infection. Fish Shellfish Immunol 2011;31:260–8.10.1016/j.fsi.2011.05.011Suche in Google Scholar PubMed
30. Alves RN, Cordeiro O, Silva TS, Richard N, de Vareilles M, Marino G, et al. Metabolic molecular indicators of chronic stress in gilthead seabream (Sparus aurata) using comparative proteomics. Aquaculture 2010;299:57–66.10.1016/j.aquaculture.2009.11.014Suche in Google Scholar
31. Chen Z, Cheng CH, Zhang J, Cao L, Chen L, Zhou L, et al. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proc Natl Acad Sci USA 2008;105:12944–9.10.1073/pnas.0802432105Suche in Google Scholar PubMed PubMed Central
32. Emmanouil C, Sheehan TM, Chipman JK. Macromolecule oxidation and DNA repair in mussel (Mytilus edulis L.) gill following exposure to Cd and Cr(Vi). Aquat Toxicol 2007;82:27–35.10.1016/j.aquatox.2007.01.009Suche in Google Scholar
33. Pruski AM, Dixon DR. Effects of cadmium on nuclear integrity and DNA repair efficiency in the gill cells of Mytilus edulis L. Aquat Toxicol 2002;57:127–37.10.1016/S0166-445X(01)00192-8Suche in Google Scholar
34. Michel C, Vincent-Hubert F. DNA oxidation and DNA repair in gills of zebra mussels exposed to cadmium and benzo(a)pyrene. Ecotoxicology 2015;24:2009–16.10.1007/s10646-015-1536-3Suche in Google Scholar PubMed
35. Ha TT, Burwell ST, Goodwin ML, Noeker JA, Heggland SJ. Pleiotropic roles of Ca(+2)/calmodulin-dependent pathways in regulating cadmium-induced toxicity in human osteoblast-like cell lines. Toxicol Lett 2016;260:18–27.10.1016/j.toxlet.2016.08.020Suche in Google Scholar PubMed PubMed Central
36. Lévesque M, Martineau C, Jumarie C, Moreau R. Characterization of cadmium uptake and cytotoxicity in human osteoblast-like MG-63 cells. Toxicol Appl Pharmacol 2008;231:308–17.10.1016/j.taap.2008.04.016Suche in Google Scholar PubMed
37. Liu Y, Templeton DM. Cadmium activates CaMK-II and initiates CaMK-II-dependent apoptosis in mesangial cells. FEBS Lett 2007;581:1481–6.10.1016/j.febslet.2007.03.003Suche in Google Scholar PubMed
38. Putnam CD, Arvai AS, Bourne Y, Tainer JA. Active and inhibited human catalase structures: ligand and NADPH binding and catalytic mechanism. J Mol Biol 2000;296:295–309.10.1006/jmbi.1999.3458Suche in Google Scholar PubMed
©2018 Walter de Gruyter GmbH, Berlin/Boston
Artikel in diesem Heft
- Frontmatter
- Research Articles
- Betaine treatment decreased serum glucose and lipid levels, hepatic and renal oxidative stress in streptozotocin-induced diabetic rats
- Oxidative stress and response of antioxidant system in Nostoc muscorum exposed to different forms of Zinc
- An investigation on Trakya region Oak (Quercus spp.) honeys of Turkey: their physico-chemical, antioxidant and phenolic compounds properties
- Effects of Shiranuhi flower extracts and fractions on lipopolysaccharide-induced inflammatory responses in murine RAW 264.7 cells
- Evaluation of resveratrol organogels prepared by micro-irradiation: fibroblast proliferation through in vitro wound healing
- Ecological and phytochemical attributes of endemic Ferula gummosa Boiss. at vegetative and generative stages
- Characterization of calmodulin in the clam Anodonta woodiana: differential expressions in response to environmental Ca2+ and Cd2+
- Foliar application effects of salicylic acid and jasmonic acid on the essential oil composition of Salvia officinalis
- Antioxidant and antimicrobial activities of four Astragalus species growing wild in Turkey
- Differences in structure, allergenic protein content and pectate lyase enzyme activity of some Cupressaceae pollen
- In vitro bioactivities and subacute toxicity study of O. basilicum, T. vulgaris and R. officinalis
- Wendlandia exserta: a pertinent source of antioxidant and antimicrobial agent
- Effects of epigallocatechin-3-gallate (EGCG) on a scleroderma model of fibrosis
Artikel in diesem Heft
- Frontmatter
- Research Articles
- Betaine treatment decreased serum glucose and lipid levels, hepatic and renal oxidative stress in streptozotocin-induced diabetic rats
- Oxidative stress and response of antioxidant system in Nostoc muscorum exposed to different forms of Zinc
- An investigation on Trakya region Oak (Quercus spp.) honeys of Turkey: their physico-chemical, antioxidant and phenolic compounds properties
- Effects of Shiranuhi flower extracts and fractions on lipopolysaccharide-induced inflammatory responses in murine RAW 264.7 cells
- Evaluation of resveratrol organogels prepared by micro-irradiation: fibroblast proliferation through in vitro wound healing
- Ecological and phytochemical attributes of endemic Ferula gummosa Boiss. at vegetative and generative stages
- Characterization of calmodulin in the clam Anodonta woodiana: differential expressions in response to environmental Ca2+ and Cd2+
- Foliar application effects of salicylic acid and jasmonic acid on the essential oil composition of Salvia officinalis
- Antioxidant and antimicrobial activities of four Astragalus species growing wild in Turkey
- Differences in structure, allergenic protein content and pectate lyase enzyme activity of some Cupressaceae pollen
- In vitro bioactivities and subacute toxicity study of O. basilicum, T. vulgaris and R. officinalis
- Wendlandia exserta: a pertinent source of antioxidant and antimicrobial agent
- Effects of epigallocatechin-3-gallate (EGCG) on a scleroderma model of fibrosis


