Abstract
In the field of biomedical research, organoids represent a remarkable advancement that has the potential to revolutionize our approach to studying human diseases even before clinical trials. Organoids are essentially miniature 3D models of specific organs or tissues, enabling scientists to investigate the causes of diseases, test new drugs, and explore personalized medicine within a controlled laboratory setting. Over the past decade, organoid technology has made substantial progress, allowing researchers to create highly detailed environments that closely mimic the human body. These organoids can be generated from various sources, including pluripotent stem cells, specialized tissue cells, and tumor tissue cells. This versatility enables scientists to replicate a wide range of diseases affecting different organ systems, effectively creating disease replicas in a laboratory dish. This exciting capability has provided us with unprecedented insights into the progression of diseases and how we can develop improved treatments. In this paper, we will provide an overview of the progress made in utilizing organoids as preclinical models, aiding our understanding and providing a more effective approach to addressing various human diseases.
Introduction
Over the past half-century, there has been remarkable progress in the field of stem cell research. In 1961, Professor Ernest A. McCulloch made a groundbreaking discovery, finding that cells from the bone marrow of mice could differentiate into various cell types. He coined the term “pluripotent stem cells (PSCs)” [1]. Professors Rheinwald and Green described the first successful serial cultivation of human epidermal cells [2]. It wasn’t until 1981 that scientists successfully isolated karyotypically normal embryonic stem cells from the inner cell mass of mouse blastocysts for the first time [3] and co-cultivated them in vitro with mitotically arrested fibroblasts. This co-cultivation was hypothesized to provide essential trophic factors for maintaining embryonic stem cell pluripotency [4]. In 1998, James Thomson and his colleagues established an embryonic stem cell line from human in vitro fertilized blastocysts that could be stably cultured in vitro with factors maintaining self-renewal and inhibiting differentiation [5]. Stem cells are known for their self-renewal and differentiation potential, allowing them to develop into specific cell types under suitable environmental conditions or through artificial induction [6]. For example, in 1980, Green and his colleagues successfully used autologous keratinocyte sheets to treat two patients with third-degree burns at the Peter Bent Brigham Hospital [7]. These advancements collectively have fostered the idea of using human stem cells to culture three-dimensional (3D) organs, holding great promise for disease treatment and drug screening.
In 1907, Professor Wilson HV achieved the first in vitro regeneration of isolated sponge cells into complete organisms through self-organization [8] (Figure 1). Subsequently, scientists generated organ-like tissues from isolated chicken embryos through dissociation-reaggregation experiments [9]. In 1978, Li et al. found that mammary epithelial cells cultured on EHS substrates can form 3D ducts and lumens capable of synthesizing and secreting milk proteins, compared to two-dimensional (2D) cultures [10]. In 2009, scientists used adult stem cells for the first time to generate 3D gut-like organs through self-organization in a matrix [11]. Organoids can be derived from both embryonic stem cells and adult stem cells [11, 12]. Milestone achievements in somatic cell reprogramming technology have significantly enriched the sources of stem cells and organoids [13], facilitating the establishment of in vitro 3D organ processes. In recent decades, the field of organoids has made impressive strides. The term “organoid” commonly refers to different 3D tissues in vitro culture, ranging from cell aggregation to organ-on-chip systems [14]. Organoids represent complex 3D tissues that mimic the structure and function of organs in vivo and are derived from stem cells specifically differentiated through a self-organizing process [15].

Timeline for the development of different types of organoids. Summary of research on critical landmarks in the establishment of various organoids. 3D, 3-dimensional; ASC, adult stem cells; hPSC, human pluripotent stem cells; mESC, embryonic stem cells.
In the past few decades, the field of organoids has made remarkable progress (Tables 1 and 2). As an emerging research platform, organoids have successfully addressed the limitations of both 2D cell culture and animal models [16]. Conventional monolayer cell cultures lack essential cellular interactions, organization, and complexity, and the immortalization of 2D cells can result in significant genetic alterations. While genetically engineered animal models have been widely used for disease modeling, they cannot fully replicate the genetic background and physiological conditions of humans. Human-derived organoids have the capacity to faithfully replicate the intricate pathological and physiological processes of in vivo organs. This capability holds great promise for applications in organ development, disease modeling, precision medicine, and drug discovery. In this context, we will describe the current development status of adult cells, stem cells, and tumor-derived organoids, and discuss their diverse applications in basic biology and preclinical research, covering the latest advances in organoid research. Finally, we will emphasize the key challenges that organoids face and their potential prospects for various applications.
Summary of organoid models.
| Organoid models | Organoid source | Species | Applications | References |
|---|---|---|---|---|
| Intestine | AdSCs | Human | Modeling of SARS-CoV-2 infection, drug screening | [37, 149] |
| iPSCs | Human | Drug screening, toxicology research | [146, 147, 151] | |
| Liver | iPSCs | Human | Gene editing, transplantation, drug screening, modeling of HBV infection | [194, 236] |
| ESCs | Human | Drug screening, personalized treatment, modeling of alcohol-related liver disease | [141, 142, 237] | |
| AdSCs | Human or mouse | Drug screening, transplantation modeling of liver cancer | [238] | |
| Lung | AdSCs | Mouse | Modeling of adenocarcinoma of lung, virus pathology, drug screening | [49, 56, 185] |
| ESCs | Human | Modeling of lung differentiation and disease | [156] | |
| iPSCs | Human | Drug screening, transplantation | [159] | |
| Heart | ESCs | Mouse | Regenerative medicine | [62] |
| iPSCs | Human | Gene editing, modeling of disease | [63, 64] | |
| Brain | ESCs | Human | Modeling of disease, gene editing; single-cell sequencing, assembly technology, vascularization technology | [67, 75], [76], [77], [78] |
| iPSCs | Human | Modeling of microcephaly, Parkinson’s disease and amyotrophic lateral sclerosis disease | [92, 123, 124, 131] | |
| AdSCs | Human | Modeling of Alzheimer’s disease | [113, 114] | |
| Kidney | Npcs | Human or mouse | Gene editing, modeling of disease, drug screening | [239] |
| iPSC | Human or mouse | Transplantation | [240] | |
| ESCs | Human | Toxicology research, transplantation | [241] | |
| Pancreas | AdSCs | Human | Modeling of pancreatic cancer, drug screening, transplantation | [43, 242, 243] |
| Retina | iPSC | Mouse | Modeling of advanced retinal degenerative disease | [163] |
| ESCs | Human or mouse | Transplantation | [25, 244] | |
| Breast | AdSCs | Human | Drug screening, personalized treatment | [198, 201] |
| Inner ear | ESCs | Human or mouse | Modeling of disease | [169, 170] |
-
AdSCs, adult stem cells; iPSCs, induced pluripotent stem cells; ESCs, embryonic stem cells; HBV, hepatitis B virus.
Summary of media compositions for different organoids culture.
| Organoid culture | Small molecule compounds | Cytokines |
|---|---|---|
| Cerebral organoids | Y-27632, MK-2206, GDC-0068, dorsomorphin | FGF-basic, Noggin, DKK-1, EGF, BDNF, GDNF, R-Spondin 1 |
| Intestinal organoids | Y-27632, SB-202190, A 83-01, gastrin, nicotinamide | EGF, Noggin, R-Spondin 1, Wnt-3a, FBS |
| Liver organoids | Y-27632, A 83-01, DAPT, forskolin, gastrin, nicotinamide, prostaglandin E2 | BMP-4, EGF, FGF-basic, FGF-10, HGF, Noggin, Wnt-3a, FBS |
| Pancreas organoids | Gastrin I, A 83-01, nicotinamide | FGF-10, EGF, Noggin, R-Spondin 1, Wnt-3a, B27 |
| Prostate organoids | Y-27632, SB-202190, A 83-01, nicotinamide, prostaglandin E2, testosterone | EGF, activin A, FGF-basic, FGF-10, Noggin, R-Spondin 1, Wnt-10b |
| Lung organoids | CHIR-99021, SB-431542 | Activin A, FGF-basic, FGF-4, Noggin, N2 |
| Mammary organoids | Y-27632, phosphorylethanolamine, isoproterenol, hydrocortisone | Here gulin β-1, R-Spondin 1, R-Spondin 2, Noggin, EGF, FGF-basic, FGF-10, Wnt-3a, prolactin |
| Kidney organoids | CHIR-99021, retinoic acid, hesparin | BMP-2, BMP-4, BMP-7, FGF-basic, FGF-9 |
-
FGF, fibroblast growth factor; EGF, epidermal growth factor; HGF, hepatocyte growth factor; BMP, bone morphogenetic protein.
Advances in organoid culture environments
Most organoids require a matrix gel to support the formation of 3D structures during development, and the most traditional and commonly used substrate is Matrigel, also called Cultrex or Engelbreth-Holm-Swarm (EHS) matrix. Matrigel, an EHS mouse sarcoma extract [17], contains laminin and a number of growth factors, which are extensively used in organoid cultures, such as gastric [18], mammary glands [19], intestinal [11], and brain [20]. Due to the uncertainty of the compositions of Matrigel and the limitations of clinical applications, more types of matrix gels were subsequently developed. One approach was to produce matrix gel by tissue decellularization. It was demonstrated that hydrogel substrates fabricated from decellularized porcine small intestine could provide better support for the growth and expansion of endoderm-derived organoids, overcoming the limitations of Matrigel tumor sources [21]. Residual animal protein constituents in matrix gels may trigger an immune response in the host, whereas synthetic macromolecular polymer matrix gels prevent this problem. Sorrentino et al. used PEG as a skeleton to integrate key extracellular matrix (ECM) proteins found in the liver, such as laminin-111, type IV collagen, and fibronectin, to create a fully chemically synthesized matrix. It was successfully employed for liver organoid culture [22]. Curvello and his colleagues developed a plant-derived matrix gel, 0.1 % plant nanocellulose fibers, which successfully cultured small intestinal organoids [23].
Cultures of nervous system-like organoids like cerebrum [20], hippocampus [24], and optic cup [25] are mostly cultured in suspension, allowing the stem cells to self-organize into 3D cell aggregates in liquid suspension. Recently, suspension culture methods have also been used for the development of other types of organoids. Capeling and her colleagues found that simple suspension cultures promoted the development of serosal mesothelial in human intestinal organoids. A comparison of suspension cultured organoids, human tissue and matrix gel cultured organoids revealed that suspension cultured organoids were more similar to human tissue at the molecular level.
Organoid chips are microfabricated cell culture devices that mimic the functional units of human organs in vitro [26]. Recent studies have demonstrated that the use of organoid-on-a-chips to construct and simulate human tissue microenvironments can recapitulate the specific physiological functions in some organs [27], [28], [29], and has a wide range of applications in the fields of organ development, regenerative medicine, drug screening, and disease modeling in the future.
Modeling human disease with organoids
Organoids derived from adult stem cells
There are two primary sources of organoids: adult stem cells (AdSCs) and PSCs, including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs). Organoids derived from AdSCs are obtained directly from human or mouse adult tissues and are further recapitulated within a tissue-like environment by suspending them in ECM and supplementing them with growth factors. In comparison to organoids derived from PSCs, AdSCs-derived organoids demonstrate superior performance in mimicking physiological and pathological conditions [30, 31].
Adult stem cells derived small intestine organoids
The first AdSC-derived organoids were developed by Hans Clevers’ research group. They established the first long-term in vitro culture of single crypts in Matrigel and maintained the crypts’ structure by supplementing them with epidermal growth factor (EGF) and R-Spondin1 [32]. These generated organoids consisted of over 40 crypt domains surrounding a central lumen lined with a villus-like epithelium, referred to as the ‘villus domain.’ This crypt structure was maintained for a remarkable 8 months. Moreover, they successfully identified and isolated the specific population of AdSCs capable of retaining their stem cell properties and forming large crypt organoids. Lgr5+ cells were found to retain these stem cell properties, and individual Lgr5-expressing stem cells could spontaneously create self-organizing crypt-villus organoids in the absence of non-epithelial niche cells [11]. Lgr5 crypt stem cells exhibited the capacity to undergo thousands of cell divisions, and a ‘mini-guts’ culture system was established, involving both whole crypts and single Lgr5+ stem cells. This system utilized a serum-free medium supplemented with three recombinant proteins: R-Spondin1 (a Wnt signal amplifier and a ligand for Lgr5), EGF, and the bone morphogenetic protein (BMP) inhibitor Noggin [33].
In contrast to mouse intestinal crypts, which are equipped with Paneth cells capable of secreting some niche growth factors [32], human gut organoid cultures lack fully functional Wnt-secreting niche cells and necessitate the addition of exogenous Wnt3a to the culture medium [34]. For the sustained maintenance of human gut organoids through multiple passages, both a TGFβ pathway inhibitor and a p38 MAPK pathway inhibitor are essential. However, the removal of the p38 MAPK inhibitor is a prerequisite for the emergence of secretory cells [35]. These inhibitors were subsequently replaced by insulin-like growth factor 1 (IGF-1) and fibroblast growth factor 2 (FGF-2). Organoids faithfully recapitulate the characteristics of their respective tissues compared to traditional 2D culture, offering significant potential in drug discovery, the study of the tumor microenvironment, exploration of tumor heterogeneity, predictive modeling of responses, and personalized medicine [31, 36].
Furthermore, the intestine, often referred to as the “second immune system”, plays a substantial role in microbiome infections. Intestinal organoids have proven invaluable in understanding the pathogenesis of colorectal cancer caused by bacterial agents. In addition to the lungs’ susceptibility to SARS-CoV-2 infection, human intestinal organoids can also be infected by SARS-CoV-2, leading to the production of substantial quantities of infectious viral particles within the intestinal environment [37].
Adult stem cells derived hepatic and biliary organoids
Hepatic and biliary (HPB) organoids can be generated through step-wise methods using either PSCs or iPSCs. Additionally, these organoids can be generated from adult human or mouse tissues. These HPB organoids are identified based on their specific tissue source, and their in vitro cell types closely mirror the guidelines outlined by Hans Clevers [38]. Hepatic organoids typically consist of cells or tissues derived from hepatocytes and cholangiocytes. LGR5, recognized as a marker for intestinal stem cells, has also been identified as a stem cell marker in various tissues. In the context of carbon tetrachloride (CCl4)-induced injury, Lgr5+ cells emerge in proximity to bile ducts. Furthermore, individual Lgr5+ cells from the damaged mouse liver can be clonally expanded as organoids when cultured in an Rspo1-based medium over an extended period. These cultured organoids, under the aforementioned conditions, express multiple markers associated with progenitor, hepatocyte, and cholangiocyte characteristics, suggesting a bipotential nature [39].
For the purpose of inducing hepatocyte differentiation, the culture medium was modified by eliminating R-Spondin and FSK, and substituting them with the Notch inhibitor DAPT, FGF19, dexamethasone, and BMP7. This alteration facilitated the expression of hepatocyte markers [40]. In 2018, Huch and colleagues developed a protocol for the long-term culture of mouse hepatocytes and human fetal liver cells as organoids. This protocol marked a departure from previous methods as it exhibited a morphological resemblance to cholangiocyte organoids. To achieve this, they employed small-molecule inhibitors, including Wnt agonists like R-Spondin1 and CHIR99021, the TGF-b inhibitor A83-01, and growth factors such as EGF, FGF7, FGF10, and hepatocyte growth factor (HGF).
The mouse Hepatocyte Organoids (mHOs), generated through this protocol, displayed a characteristic ‘bunch-of-grapes’ structure. These organoids expressed specific hepatocyte markers, including Alb, Hnf4a, Cyp1a2, Cyp3a11, and the fetal hepatocyte marker Afp. Importantly, they did not exhibit markers associated with cholangiocytes or progenitor cells, such as Krt19, Tbx3, and Sox9. Additionally, these mHOs bore a resemblance to hepatocytes observed after partial hepatectomy [41].
Hepatocyte organoids, aside from Wnt-mediated cultures, have been successfully established using an inflammatory cytokine-mediated expansion protocol for long-term cultured mHOs. In the context of liver regeneration, tumor necrosis factor-alpha (TNF-α) activates a series of transcription factors, including nuclear factor kappa B (NF-κB), Janus kinase (JAK/STAT), activator protein 1 (AP-1), and Yes-associated protein (YAP), which enhance cell proliferation. This culture condition presents an innovative approach to expanding primary mouse hepatocytes in 3D culture [41]. Under these expansion conditions, mHOs could be sustained for at least 8 months and exhibited similarities to proliferating hepatocytes following partial hepatectomy (PHx) [42]. This study introduces a novel approach to establishing hepatocyte organoids. The hepatocyte organoids culture system could be extended to propagate primary liver cancer (PLC) organoids, covering three of the most prevalent PLC subtypes: hepatocellular carcinoma (HCC), cholangiocarcinoma (CC), and combined HCC/CC (CHC) tumors [43, 44].
Adult stem cells derived lung organoids
Lungs are complex structures characterized by a branching network of airways and blood vessels, resembling a tree-like configuration. This intricate system begins with the trachea and extends into branching airway tubes, ultimately culminating in millions of air sacs known as alveoli where gas exchange with the vasculature takes place [45, 46]. The most proximal airway, the trachea, bifurcates at the carina, giving rise to the left and right main stem bronchi. Each main bronchus further subdivides into secondary or lobar bronchi, which subsequently branch into progressively narrower airways until the smallest bronchioles connect to the alveoli [47]. Various cell types are present in human lungs, encompassing epithelial cells, endothelial cells (comprising vasculature and lymphatics), pleura/mesothelium, airway and vascular smooth muscle cells, pericytes, fibroblasts, neurons, and immune cells such as alveolar macrophages [48].
Lung organoids can be generated from various cell types, including basal cells, airway secretory cells, and alveolar type II cells (AEC2 cells). Two primary approaches to obtain target cells are lineage-tracing and surface marker-based cell sorting. The initial organoids derived from mouse tracheal basal cells were referred to as “tracheospheres.” These cells were isolated from airway basal cells expressing Trp-63 (p63) and cytokeratins 5 (Krt5) and Krt14. Basal cells play a crucial role in generating differentiated cells not only during postnatal growth but also in adults under steady-state conditions and during epithelial repair [49, 50]. Organoids obtained from isolated basal cells may be classified as either tracheospheres or bronchospheres, depending on whether the basal cells originate from the trachea or the larger airways. Typically, these organoids comprise TRP63+ KRT5+ basal cells, functional multiciliated cells, and secretory goblet cells [51, 52].
Organoids derived from airway secretory cells are primarily generated by isolating secretory Club cells based on surface markers. Lung epithelial cells that fulfill specific criteria, being CD45 (Ptprc)neg, CD31 (Pecam1)neg, EpCAM high, CD49f (Itga6)pos, CD104 (Itgb4)pos, and CD24 low, are capable of forming spheres. These spheres can be categorized into three general groups based on their morphology after 14 days of culture [53]. To generate ‘alveolospheres’, a lineage-tracing approach was initially employed [54]. In the case of human AEC2s, isolation is typically achieved using a monoclonal antibody that specifically targets human AEC2s, known as HTII-280 [55]. Throughout the COVID-19 pandemic, human lung organoids have proven to be a valuable preclinical model for investigating virus pathobiology and advancing therapeutic development. SARS-CoV-2 productively infects AEC2s, leading to the initiation of an innate immune response. The infection triggers both cell-autonomous and non-cell-autonomous apoptosis, potentially contributing to alveolar injury [56, 57].
Organoids derived from pluripotent stem cells
Cardiac organoids
Cardiovascular diseases are the leading global cause of death. It has long been difficult to obtain satisfactory and ideal cardiac models for cardiovascular disease studies in vitro. Recent advances in cardiac organoids have provided access to more accurate models with complex structures and functional maturation for unveiling cardiac developmental biology, modeling cardiovascular diseases, establishing relevant drug screening platforms, and exploring transplantation therapies.
A large body of studies has provided insights into methods of constructing heart-like organs and cardiac developmental biology. In the past decade, scientists have successfully employed tissue engineering techniques to facilitate the formation of cardiac chambers and emulate organ complexity, including hydrogels and biomaterial-based scaffolds [58]. Recent studies have made significant progress in generating cardiac organoids that can simulate heart development in vivo in a spatial and temporal manner through self-organization. These organoids are powerful tools for studying cardiac development [59, 60]. The first functional ‘mini-heart’ organoids induced by mouse embryonic stem cells (mESCs) were reported in 2020 [60]. mESCs undergo a self-organized event to form heart-like structures when exposed to FGF4 and LN/ET. These structures showed considerable similarity to the developing heart in vivo, possessing four intact chambers and a conducting system that exhibited myocardial contraction and action potentials [60]. In a pioneering study published in 2021, Professor Mendjan and his group developed the first in vitro self-organizing human cardiac organoid model. This model spontaneously forms a cavity and beats on its own without the need for stent support. They found that cavity morphogenesis is governed by a mesodermal WNT-BMP signaling axis, and the cardiods can autonomously mobilize cardiac fibroblasts to migrate and repair damage after injury [59].
Cardiac organoids, which possess a 3D structure allowing them to mimic interactions between different cell types and complex pathophysiological processes, serve as an excellent platform for cardiovascular disease research. These miniature models are invaluable for investigating conditions like myocardial infarction, arrhythmia, and genetic cardiac diseases. Complementing pre-clinical models, cardiac organoids help uncover pathogenic mechanisms and detect drug toxicity and side effects. In a recent study by Richards et al., they created cardiac organoids that leverage nutrient transport principles and stimulated them with the neurotransmitter noradrenaline to mimic the post-myocardial infarction structure of the human heart in vitro. Through transcriptomic analysis and structural and functional validation, they successfully established an in vitro 3D model of myocardial tissue after myocardial infarction [61]. Voges and colleagues demonstrated that human cardiac organoids resembling fetal heart tissue could model acute myocardial infarction following cryoinjury with a dry ice probe. These cardioids exhibited an endogenous regenerative response, achieving full functional recovery within 2 weeks of acute injury. This study highlights the regenerative capacity of immature human heart tissue in response to injury [62]. In a recent study, Hofbauer et al. utilized an advanced hPSC-derived self-organizing cardiac organoid model to assess the effects of freezing injury on the heart. They observed that cardiac fibroblasts migrated toward the injury site and produced proteins to repair the damage [59]. Cardiac organoids are also utilized to study arrhythmia. Goldfracht et al. developed an arrhythmia model based on atrial engineered heart tissues and demonstrated its effectiveness by applying relevant pharmacological interventions. They confirmed the ability of the antiarrhythmic agents flecainide and vernakalant to terminate re-entrant activity in the atrial organoid model [63]. Today, the combination of gene editing technology and cardiac organoid disease models holds promise for addressing genetic heart diseases. Mutations can be precisely corrected and mitigated to provide personalized treatment for patients. For instance, Chengzu Long and his colleagues corrected Duchenne muscular dystrophy (DMD) mutations through exon skipping in 3D engineered heart muscle. This approach led to restored dystrophin expression and the corresponding mechanical force of contraction. Their work demonstrated that single-site genomic editing in just 50 % of cardiomyocytes is sufficient to rescue mutant engineered heart muscle phenotypes to near-normal control levels [64]. However, it is important to note that current cardiac organoids remain relatively simplified, lacking the tissue microenvironment found in vivo, such as the immune and nervous systems. Bridging this gap between in vivo and in vitro applications of cardiac organoids will require further efforts and advancements.
Cerebral organoids
Studying and effectively treating diseases of the nervous system is a formidable challenge, primarily due to the intricate nature of the human brain and its inaccessibility. The human brain, arguably the most complex organ in our body, comprises an extensive array of highly specialized cells that function in an interconnected manner [65]. While there have been promising advances in investigating the molecular and biological mechanisms of neurological development and neuropsychiatric disorders using monolayer neuronal cultures derived from human pluripotent stem cells and animal models, there are limitations. The former lack precise cellular connectivity across various regions of the brain, as well as many distinguishing features and species differences that hinder animal models in neurobiology research [66]. Brain organoid techniques offer a solution to the shortcomings of animal models and 2D cellular models. They provide a more accurate representation of in vivo conditions by reproducing the diversity of human neural cells, the physiological environment of nervous system development, and the intricacy of neural networks.
The brain organoid technique, first established in 2013 [20], originated in Knoblich’s laboratory. They developed a 3D organoid culture system using human pluripotent stem cells to generate cerebral organoids. These brain-like organoids have the ability to develop various discrete but interdependent brain regions. The cerebral cortical regions within these organoids faithfully recapitulate features of human cortical development, including apical-basal polarity, dorsal cortical cytoarchitecture, interkinetic nuclear migration during radial glial division, and the pattern of neural migration [20]. Cerebral organoids typically originate from pluripotent stem cells, including ESCs derived from early embryos and iPSCs obtained through somatic reprogramming. PSCs can be cultured and differentiated into various types of human cerebral organoids within 3D environments, either through the intrinsic self-organizing capacity of aggregated cells or by induction with specific developmental regulators [67]. To date, researchers have successfully established a variety of cerebral organoids, including whole-brain and specific brain region organoid systems, such as those modeling the cerebral cortex, midbrain, cerebellum, ventral telencephalon, thalamus, hypothalamus, striatum, and hippocampus [24, 68], [69], [70], [71], [72], [73]. Numerous studies have shown that cerebral organoids effectively simulate in vivo processes like neurogenesis, neuronal migration, cortical stratification, and the establishment of neural circuits. They have significantly advanced our understanding of neurodevelopmental disorders and neurodegenerative diseases in vivo [66]. Substantial progress has been made in the study of conditions such as Miller-Dieker syndrome (MDS), microcephaly-related disorders, autism spectrum disorder (ASD), tuberous sclerosis complex, brain tumors, and Alzheimer’s disease (AD), among others [74]. Notably, an increasing number of cutting-edge technologies, such as gene editing, single-cell sequencing, assembly techniques, and vascularization technologies, have been applied to cerebral organoids. This holds great promise for the advancement of research and treatment in the field of neurological disorders [75], [76], [77], [78].
Neurodevelopmental disease
Early neurodevelopmental abnormalities can lead to malformations of the cerebral cortex, giving rise to disorders such as lissencephaly, microcephaly, Timothy syndrome, and tuberous sclerosis complex, as well as neuropsychiatric disorders, including ASD and schizophrenia (SZ). Human cerebral organoids have emerged as a promising platform for the study of neurodevelopmental and psychiatric disorders.
Lissencephaly
MDS is a severe cortical malformation characterized by defects in cortical folding and radial migration. It is commonly associated with microcephaly, cognitive impairment, mental retardation, and intractable epilepsy [79, 80]. MDS is caused by multiple heterozygous deletions of human chromosome 17p13.3, including PAFAH1B1 (LIS1 protein) and YWHAE (14-3-3ε protein) [81]. In research published in 2017, both Bershteyn et al. and Iefremova et al. developed forebrain organoids to deepen our understanding of MDS. Studying MDS in animal models is challenging because mice naturally have lissencephalic brains [82, 83]. Outer radial glia (oRG) progenitors, known for their robust proliferation potential, produce large numbers of neurons and offer divergent tracks for neurons migrating along their basal fibers, facilitating cortical folding [84, 85]. When compared with control organoids, hiPS-derived telencephalic organoids from MDS patients displayed increased apoptosis, horizontal division of neural stem cells, and impaired neuron migration. Moreover, prolonged mitosis of oRG suggests that oRG may play a role in the pathogenesis of lissencephaly [82]. Additionally, the N-cadherin/beta-catenin signaling axis was impaired, resulting in defective non-cell-autonomous expansion of radial glial cells in the ventricular zone. Pharmacological activation of Wnt signaling ameliorated the abnormal growth of MDS patient-derived organoids [83].
Microcephaly
Microcephaly is a neurodevelopmental disorder characterized by abnormal brain development, resulting in a significant reduction in brain volume in affected individuals [86]. Gene mutations can lead to primary microcephaly, and extensive studies have connected several genes with autosomal recessive primary microcephaly (MCPH), such as CDK5RAP2, ASPM, NARS1, and IER3IP1 [87], [88], [89]. The first cerebral organoid was established to study microcephaly. Mutations in the CDK5RAP2 gene, which encodes centromere protein, can lead to autosomal recessive primary microcephaly in humans [90]. When cerebral organoids were generated from MCPH patients carrying heterozygous mutations in the CDK5RAP2 gene, they exhibited premature neuronal differentiation at the expense of the progenitor cell pools, resulting in smaller-sized brain organoids. Overexpression of the CDK5RAP2 gene could rescue the reduced size of cerebral organoids, and the phenotype could be reproduced by RNAi-mediated knockdown of CDK5RAP2 [20]. The most common cause of MCPH is mutations in the ASPM gene, which are critical for mitotic spindle function [85]. Cerebral organoids with biallelic mutations in ASPM exhibited less neuroepithelial organization, fewer vRG and oRG cells, and defective layer lamination, indicating developmental defects in the early stages. In later stages of development, calcium imaging experiments revealed fewer mature neurons and less synchronization of neuronal activities in organoids with ASPM mutations [91]. iPSC-derived cortical brain organoids with NARS1 mutations also displayed a substantial reduction in organoid size, linked to a decrease in neural rosette size and neuron number after mitosis, which may elucidate the pathogenesis of microcephaly [92]. To date, cerebral organoids have been employed to screen for genes contributing to microcephaly. It was found that IER3IP1 could modulate brain growth by regulating the unfolded protein response [93].
In addition to primary microcephaly, scientists have harnessed brain organoids to understand microcephaly caused by pathogens. It has been discovered that cerebral organoids show reduced proliferation of neural progenitor cells and undergo cell death mediated by cysteine asparaginase when exposed to the ZIKV virus [94]. The NS2A protein encoded by the ZIKV virus disrupts the formation of adhesion junctions and impairs the proliferation of radial glial cells in human forebrain organoids [95]. In human fetal neural stem cells infected with ZIKV, two other proteins, NS4A and NS4B, synergistically inhibit the Akt-mTOR signaling pathway, blocking neurogenesis and leading to abnormal autophagy. This may provide insights into the pathogenesis of ZIKV virus-induced microcephaly [96]. Furthermore, organoid models of ZIKV virus-induced microcephaly have been used to test and screen potential therapeutic drugs [97, 98].
Autism spectrum disorder
ASD refers to a heterogeneous neurodevelopmental disorder caused by mutations in multiple genes [99]. ASD patients typically exhibit childhood-onset symptoms, including repetitive behaviors, restricted interests, and impaired social interactions and communication [74]. Understanding the pathophysiology of ASD has been challenging due to the extreme complexity of its pathogenesis, which involves interactions with multiple inherited factors and environmental influences.
Emerging evidence demonstrates that cerebral organoids are excellent tools for unraveling the mysteries of ASD. In a study published in 2015, scientists developed telencephalic organoids derived from iPSCs from severe idiopathic ASD patients and family controls [100]. Transcriptome and gene network analyses of ASD-derived organoids revealed upregulated genes associated with cell proliferation, neuronal differentiation, and synaptic assembly. Compared to control organoids, they found that the cell cycle was accelerated, and GABAergic inhibitory neurons were overproduced in ASD-derived organoids. Significantly, aberrant upregulation of the transcription factor FOXG1 was identified as responsible for the overproduction of GABAergic neurons through RNA interference experiments, and knockdown of FOXG1 rescued this phenotype [100]. In another study, CHD8, which encodes a chromatin remodeling factor, was found to affect GABAergic neuron development in ASD-derived telencephalic organoids by modulating DLX gene expression [101]. More recently, a pioneering study explained how different risk genes consistently contribute to the characterization of ASD phenotypes in human cerebral cortex organoid models [102]. Evidence suggests that mutations in three ASD risk genes – SUV420H1, ARID1B, and CHD8 – can lead to asynchronous development of two major cortical neuronal lineages: γ-aminobutyric acid-releasing (GABAergic) neurons and deep-layer excitatory projection neurons. The expressivity of these mutations is closely regulated by individual genomic context, suggesting the pathological basis for the varying clinical manifestations of ASD patients and emphasizing the importance of future ASD treatment focusing on common pathways influenced by ASD risk genes [102].
Neurodegenerative disorders
Neurodegenerative diseases (NDDs) constitute a group of neurological disorders characterized by the progressive loss or degeneration of neurons in the nervous system, leading to memory, sensory, cognitive, and motor dysfunction [103, 104]. Some well-known NDDs include AD, Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and Huntington’s disease (HD). Despite substantial efforts to understand the pathogenesis of NDDs, much remains unknown. Major pathological mechanisms include pathological protein aggregation and autophagy defects, aberrant mitochondrial energy metabolism, neuronal cell death, neuroinflammation, and dysfunction of synapses and neuronal networks [103]. There is an urgent need for effective therapeutic approaches and drugs for NDDs. Human cerebral organoids can significantly advance NDD research by overcoming the limitations of animal models, which possess different genetic backgrounds from humans, and simple 2D cell cultures that lack multiple cell types needed to replicate the in vivo environment.
Alzheimer’s disease
AD is a neurodegenerative disease characterized by its insidious onset and progressive loss of higher neurological functions [105]. A hallmark pathology of AD is the deposition of neuronal extracellular amyloid-beta (Aβ) and the formation of neurofibrillary tangles, which consist of intracellular aggregates of hyperphosphorylated tau (Phosphorylated Tau, P-Tau) protein [106, 107]. In 2016, the first cerebral organoid derived from patients with familial AD carrying a repeat mutation in the APP gene was established [108]. Subsequently, familial AD brain organoid models with mutations in PSEN1, PSEN2, and MAPT genes, as well as sporadic AD brain organoids, have also been reported [109], [110], [111]. All of these models successfully recapitulate the pathological features of AD, including amyloid aggregation and abnormal hyperphosphorylated tau protein. In a study published in 2018, Joseph et al. introduced a new 3D human AD triculture model. This model incorporates neurons, astrocytes, and microglia within a 3D microfluidic platform to facilitate microglia recruitment and the secretion of pro-inflammatory cytokines and chemokines, providing a more accurate organoid model of the human brain [112].
Human brain organoids derived from AD patients offer profound benefits for advancing mechanistic studies and therapeutic treatments. The APOE gene stands as a major risk factor for AD. Scientists have discovered that APOE4 astrocytes increase the formation of neuronal lipid droplets and cholesterol accumulation in chimeric organoids of astrocytes and neurons, promoting the onset of AD [113]. The co-occurrence of APOE4 astrocytes and neurons is essential for elevating neuronal tau protein levels, underscoring the significance of both neuronal and astrocytic APOE4 in p-tau pathology [113]. Pérez et al. constructed mitochondrial protease 1 (PITRM1) knockout cerebral organoids and observed that the early activation of the mitochondrial unfolded protein response (UPRmt) led to mitochondrial dysfunction, resulting in AD-related phenotypes [114]. There is evidence suggesting that isogenic conversion of the AD-causing gene APOE4 to APOE3 attenuates APOE4-related phenotypes in brain organoids from AD patients [115]. Furthermore, AD cerebral organoids also serve as an effective platform for the development and screening of therapeutic drugs for AD patients. The histone deacetylase 6 (HDAC6) inhibitor CKD-504 can degrade pathological tau proteins in cerebral organoids and ameliorate the pathological phenotype of AD [116]. Park et al. have developed a network-based drug-screening platform by integrating mathematical modeling and pathological characterization of organoids from AD patients, aiming to provide a strategy for precision medicine [117].
Parkinson’s disease
PD, the second-most prevalent neurodegenerative disease globally, has become the fastest-growing neurological disease in terms of prevalence, disability, and mortality [118]. Similar to AD, the definitive cause of PD remains unclear, with research in recent years suggesting connections to genetics, the environment, and aging factors [119]. Among its pathologies, progressive loss of midbrain substantia nigra dopaminergic neurons and the accumulation of intracytoplasmic Lewy Bodies, eosinophilic inclusions, lead to the severe symptoms of PD [119, 120].
In 2016, Jo et al. cultured midbrain organoids containing neurons of distinct layers and detected electrically active and functionally mature midbrain dopaminergic neurons (mDAN) with the ability to produce dopamine [72]. In recent years, scientists have successfully replicated the typical pathological features of PD in midbrain organoids harboring different genes associated with PD. In iPSC-derived midbrain organoids from PD patients carrying a triplication of the SNCA gene, increased expression levels of α-synuclein, a major component of Lewy bodies, were observed, and the accumulation of alpha-synuclein over time was linked to a selective loss of dopaminergic neurons [121]. Furthermore, midbrain organoids carrying mutations in the LRRK2 gene from PD patients exhibited neurodevelopmental defects, resulting in a decreased number of mDAN compared to controls [122]. Additionally, the induction of gene-edited iPSCs into midbrain organoids recapitulated hallmark features of PD [123, 124], underscoring the utility of established disease models for PD research.
Midbrain organoids derived from PD patients are promising models for therapeutic research and drug screening. Midbrain organoids with LRRK2 mutations from PD patients displayed increased LRRK2 activity, reduced dopaminergic neurons, and higher levels of autophagy [122]. Treatment with the LRRK2 inhibitor PFE-360 rescued the phenotype by increasing dopamine release and improving the function of dopaminergic neurons [122]. In another study, treatment with 2-hydroxypropyl-β-cyclodextrin ameliorated neuronal autophagy and mitophagy capacity in PD patient-specific midbrain organoids, leading to an increase in neuronal dopaminergic differentiation [125].
Amyotrophic lateral sclerosis
ALS is a neurodegenerative disease characterized by progressive muscle weakness due to the death of motor neurons in the brain, brainstem, and spinal cord [126]. Clinical symptoms of ALS patients include progressive muscle atrophy and cognitive decline, with ALS patients typically succumbing to respiratory failure within 5 years [127]. The etiology of ALS is not yet fully understood. Approximately 15 % of ALS cases are familial ALS (fALS), while the remainder are sporadic ALS (sALS) [128]. To date, approximately 40 genes linked to ALS have been identified, primarily influencing protein homeostasis, DNA damage repair, RNA metabolism, vesicle transport, and mitochondrial function [129]. Some of these genes are also associated with other neurological disorders, such as CCNF, ANXA11, TBK1, and SQSTM1, which are related to frontotemporal dementia (FTD) [129].
Scientists have harnessed the potential of human cerebral organoids to delve into the pathogenesis and treatment of ALS. For instance, Szebényi et al. developed a cerebral organoid slice model derived from human iPSCs, displaying early molecular pathology of C9ORF72 ALS/FTD [130]. Combined with single-cell sequencing, they observed protein disorders in astrocytes, including the early accumulation of the autophagy signaling protein P62 and the toxic dipeptide repeat protein poly (GA), along with cell death occurring in deep layer neurons. Furthermore, GSK2606414, a repressor of translational inhibition caused by UPR, could pharmacologically rescue the phenotype [130]. Researchers have also recently generated microglia-containing cerebral organoids derived from iPSCs from ALS patients [131]. Impaired microglia-mediated autophagy and down-regulated expression of the type I interferon signaling pathway were detected in patient-derived cerebral organoids compared to controls, suggesting that microglia are also involved in the pathogenesis of ALS [131]. Additionally, Pereira et al. have generated sensory-motor organoids with physiological neuromuscular junctions (NMJs) and applied this model to different subgroups of ALS, contributing to a better understanding of the pathophysiological mechanisms of ALS [132].
Liver organoids
The liver, one of the essential organs in the human body, performs various functions, including metabolism, synthesis, detoxification, and bile secretion [133]. However, liver disease remains a significant global public health concern. According to the World Health Organization’s statistics, 350 million people worldwide suffer from liver disease, with more than 1 million people succumbing to it annually. The primary causes are the scarcity of organ donors and an incomplete understanding of the pathological mechanisms [134]. Therefore, the study of liver development, regeneration, and pathogenesis holds immense significance in regenerative medicine and disease treatment.
Traditional methods for studying liver diseases have their limitations. For instance, in a 2D cell culture system, isolated primary human hepatocytes struggle to maintain viability over an extended period in vitro and have limited expansion capabilities. This system is suitable only for short-term studies, and the cells’ functions deteriorate rapidly after 48 h. Conventional animal models may not be applicable to all diseases, with variations among species and potential ethical constraints [135, 136]. The development of liver organoids offers a more physiologically relevant and in vivo-like environment for studying liver diseases [137].
The induction of iPSCs toward mesoderm or endoderm, particularly for hepatocyte and cholangiocyte differentiation, involves the use of specific cytokines such as fibroblast growth factor (FGF), BMP, HGF, and Wnt. Liver organoids are subsequently formed by embedding these cells in Matrigel under the influence of specific factors. Another approach for liver organoid formation, pioneered by Takebe and colleagues, involves the spontaneous formation of 3D aggregates known as iPSC-liver buds (iPSC-LB). These liver buds are created by mixing human pluripotent stem cell-derived hepatocytes, human mesenchymal stem cells, and human endothelial cells on a Matrigel layer, resulting in an organ-like in vitro liver model with functional vascular-like endothelial networks. Subsequent experiments involving the implantation of liver buds into mouse models of liver disease have demonstrated their potential for reducing mortality in mice with liver injuries [138]. Studies have shown that hPSC-derived liver organoids not only secrete human albumin and alpha-1-antitrypsin (A1AT) but also synthesize urea and regulate cytochrome P450 (CYP) enzymes in vitro.
Liver organoids have become invaluable tools in various therapeutic fields. For instance, HCC often results from chronic parenchymal liver damage due to monogenic diseases. Liver organoid models are indispensable for studying this disease [139]. It has been reported that hepatocyte-derived organoids, derived from iPSC-derived cholangiocytes in 3D culture, can excrete bile acids and exhibit functional secretion, mimicking conditions such as Alagille syndrome [140]. iPSC-derived liver organoid models of steatohepatitis hold the promise of providing personalized drug treatment options for reducing lipid accumulation, potentially mitigating the impact of Wolman’s disease on patients [141]. Additionally, iPSC-derived liver organoids from patients with specific diseases can simulate various inherited metabolic disorders and cholangiopathies, offering a platform for in vitro drug validation for conditions like polycystic liver disease and cystic fibrosis cholangiopathy [142].
In the past decade, hepatocytes and liver organoids derived from iPSCs have played a significant role in the in vitro study of liver physiology and pathology. However, challenges related to limitations, safety, and stability still exist. Efforts are ongoing to enable their clinical application and help address various disease-related issues as soon as possible.
Intestine organoids
Hans Clevers’ laboratory has initiated a new chapter in the development of intestinal organoids, utilizing single mouse LGR5 intestinal stem cells to self-organize into an intestinal crypt-villus structure in vitro. In the past, biological model systems such as the mouse model were employed to study enteroendocrine cells. Additionally, certain cell lines have been used as research models, such as the mouse GLUTag cell line, which serves as a model for regulating GLP-1 secretion [143]. There are also BON cell lines for endometrial cancer, which can be used to model serotonin production [144]. While these models have advanced the study of the enteroendocrine system to some extent, they still possess certain limitations, including the inability to simulate the in vivo environment and a lack of cell-to-cell interactions. Over the past decade, organoid models derived from AdSCs have been developed to investigate interactions between enteroendocrine cells, other epithelial cell types, and the enteric nervous system. However, the iPSC method can construct a more ideal model suitable for disease research.
For colonic diseases, studies have been conducted to create colonic organoids. Colon organoids were generated from iPSCs derived from patients with familial adenomatous polyposis (FAP). PSCs were induced into directed endoderm using CHIR and activin A. In the presence of B27, CHIR, and FGF4 were utilized to induce hindgut endoderm (HE). Starting from day 8, HE cells were treated with a medium containing CHIR, LDN, and EGF for 12 days to produce colonic epithelial cells, which were subsequently embedded in Matrigel to gradually form organoids [145]. Compounds that have been reported to be effective against colon cancer, such as rapamycin [146] and XAV 939 [147], can be considered as potential drugs to treat FAP. The results have shown that these two compounds reduced cell proliferation not only in colon cancer but also in normal colon tissue. However, this potential drawback, which could harm healthy colonic crypts, may limit their therapeutic use. In contrast, geneticin reduces the over-activation of WNT signaling, thereby decreasing the excessive proliferation of colonic epithelial cells, which is strongly associated with colorectal cancer. One of the advantages of disease modeling is the ability to screen for drugs that can reverse the pathogenic phenotype [145].
The main symptoms of COVID-19 primarily affect the respiratory tract, but ACE2 expression is highest in the microvilli of intestinal epithelial cells in the human body. Relevant studies have suggested that the gastrointestinal tract may serve as a potential entry route for SARS-CoV-2 [148]. Numerous experiments conducted with intestinal organoids have demonstrated that iPSC-derived intestinal organoids can be employed as a research model to replicate COVID-19 infection. For instance, remdesivir and EK1, two viral fusion inhibitors, have been discovered to inhibit coronavirus infection and restore the morphology of intestinal organoids [149]. Certain studies have utilized intestinal organoids induced by coronavirus infection and subsequently employed TEM imaging and gene set analysis to unveil the presence of viral particles and the upregulation of apoptosis-related genes.
The study of these intestinal models can further our understanding of the physiology, developmental biology, regenerative medicine, and pathophysiology of the intestine. Moreover, they can be employed to investigate host-pathogen interactions, digestion and absorption processes, transplantation, and more. For example, intestinal organoids can be used to explore intestinal barrier integrity and metabolism [150], iPSC-derived intestinal organoids that contain the enteric nervous system can help investigate intestinal motility disorders, and they can even serve as models for toxicology assessments [151].
Traditional organoids are primarily derived from homeostatic stem cells. However, when the body is damaged, homeostatic stem cells may not be sufficient to compensate for tissue loss. Previous studies have reported that constructing a proliferative intestinal organoid system can better reflect the regenerative response of the intestinal epithelium to injury in vivo, and have identified VPA and EPZ 6438 as important regulators of intestinal organoid proliferation [152]. This innovative organoid system may play a more critical role in exploring intestinal diseases and regeneration in the future. Nonetheless, iPSC-derived organoids have smaller cell populations than normal tissue and tend to decline over time [153]. The controlled formation of intestinal organoids remains a major challenge for future research [154].
Lung organoids
Previous studies have provided a strategy for deriving lung organoids from iPSCs. Initially, iPSCs are differentiated into endoderm in a medium containing activin A. Subsequently, the definitive endoderm is cultured in a medium containing NOG, FGF4, CHIR99021, and SB431542 to encourage differentiation into foregut spheroids. Finally, these foregut spheroids are embedded in Matrigel for culture, with the addition of FGF10 to the medium.
Since the development of lung organoids, extensive basic research has been conducted. For instance, NKX2.1 in lung epithelial progenitor cells can be used to screen for markers that enhance expression, allowing for more efficient and targeted differentiation towards specific fates [155]. Some experiments have revealed that the culture process of iPSC-derived lung organoids shares common features with fetal lung development. Under conditions that stimulate FGF signaling, lung organoids preferentially generate NKX2.1, SOX2, and SOX9 epithelial cells, resembling early lung progenitors [156]. When lung organoids are placed in media that activate BMP, FGF, and WNT signaling pathways, lung epithelial cells undergo morphological branching, akin to the processes during lung development [157].
Furthermore, the advent of lung organoids has facilitated the modeling and study of various diseases, including pulmonary fibrosis, congenital diseases, and neonatal respiratory distress syndrome. In the wake of the global COVID-19 pandemic, iPSC-derived lung organoids have proven valuable for modeling the disease, and candidate drugs for treatment have been tested in relevant studies [150, 158]. Another possibility is the induction of human lung lobes from hPSCs. Although the coverage of human lobes is not as extensive as in the rat model, this in vitro lung engineering approach may address the issue of the shortage of lung donors for transplantation [159].
Numerous efforts have been made to address idiopathic pulmonary fibrosis (IPF), but the pathophysiological mechanism of the disease remains poorly understood. The primary reason is the lack of models for studying pathological mechanisms and drug screening. While animal models and 2D cell cultures have contributed to progress in exploring the mechanisms of IPF, they also come with their own limitations. IPSC-derived organoids, including lung bud organoids and alveolar epithelial organoids, offer a more accurate simulation of the in vivo environment, making them a powerful model for studying respiratory diseases [160]. Nevertheless, the lung is a highly complex organ, and IPF involves the interaction of various cell types, making it a significant challenge to decipher the pathology. The development of human fetal-derived mesenchymal organoids for IPF modeling [161] and fibrotic lung organoids created through the CRISPR/Cas9 gene editing system [162] have been instrumental in exploring the pathological mechanisms of IPF.
It is undeniable that iPSC-derived lung organoids hold great potential for studying specific aspects of human development and disease. Lung organoid systems provide a valuable platform for basic research, toxicity assessment, drug screening, and personalized disease modeling.
Retinal organoids
The retina originates from the neuroectoderm and functions as a light-sensitive region of the eye. During embryonic development, the optic vesicle invaginates to form the optic cup divided into two layers, with the outermost layer forming the retinal pigment epithelium and the inner layer highly specialized to constitute the retinal neuroepithelial layer composed of retinal neurons and glial cells. The neural retina then continues to stratify, with the development of layers of photoreceptor and ancillary cell types, such as horizontal cells, bipolar cells, and so on.
Retinal morphogenesis depends on an intrinsic self-organizing program, and pluripotent stem cells can differentiate into retinal organoids by regulation of endogenous factors. Optic nerve cup-like organoids derived from mESCs can aggregate and develop in serum-free medium containing low levels of growth factors to form stratified neural retinal tissues, similar to the developmental trajectory in vivo [25]. When transplantation of ESC or iPSC-derived retinal cells into a model of advanced retinal degeneration without the outer nuclear layer, the transplanted tissue could establish synaptic connections with host cells and restore light response. The studies provided proof of concept for transplantation therapy of retinal cells [163, 164]. Three-dimensional optic nerve cup organoids of hiPSC origin can spatiotemporally recapitulate the major steps of retinal development, and differentiate into mature photoreceptors, giving rise to possibilities for disease modeling and future therapies for retinal dysfunction [165]. Recently, a human retinoblastoma model was successfully established in mice. hiPSCs from retinoblastoma patients were cultured and differentiated into retinal organoids and then transplanted into the vitreous of mouse eyes to support tumor growth, which was molecularly, cellularly and genomically identical to the tumor patients [166]. This human cancer model facilitates the study of pathogenesis and specific treatments for patients with particular germline cancers.
Inner ear organoids
The inner ear develops from the otic placode of the ectoderm. The inner ear contains a large population of specialised sensory cells, known as ‘hair cells’, which are able to convert mechanical stimuli into electrochemical signals [167]. The development of the inner ear is modulated by signaling pathways such as BMP, Wnt and Sox2, and regulating the inputs of these signaling factors in vitro induces the directed differentiation of PSCs towards sensory hair cells in the inner ear [168]. In 2013, Koehler and his colleagues partially recapitulated the developmental process of inner ear in vitro by temporally controlling the signaling pathway with precise timing and differentiating mouse ESCs into functionally mature sensory hair cells in a 3D culture system [169]. Subsequently, by a similar approach, Koehler induced inner ear organoids innervated by sensory neurons from human ESCs [170]. Recently, multiple immunostaining and single-cell RNA sequencing techniques were used to compare the characteristics of early human embryonic otocyst and fetal sensory organs with the human inner ear organoids, together with an assessment of the expression and localization of key markers at the same stage, providing evidence that the inner ear organoids can be further used as a developmental and disease models [171].
Since the first technical breakthrough of intestinal organoids in 2009, more and more organoids have been cultured and applied to various organs, including brain, liver, lung, pancreas, kidney, etc., which can reduce the complexity of experimental operation and better optimize the experimental protocol. They have important applications in drug screening, transplantation, disease modeling, gene editing, personalized treatment, and toxicology research. In terms of disease modeling, even complex and fine brain organs can be constructed to construct structures such as cerebellum and cortical layers for the study of various diseases. The kidney and liver organoids, which are the most important metabolic functions, also provide a platform for toxicity prediction. In addition to iPSCs and ESCs, organoids derived from primary tumors such as colon, breast, and pancreas can also be used for drug screening and provide substrates for personalized therapy. In summary, more studies are needed to evaluate the efficacy and safety of these methods to guide the clinical application of organoids (Table 1).
Organoids derived from tumor
Cancer is one of the significant threats to human health worldwide. In current preclinical research on tumors, there are several types of tumor models: tumor cell line models (CCL), patient-derived tumor xenograft models (PDTX), and patient-derived organoids (PDO).
CCL, originating from immortalized cancer cell lines derived from patient tumors, are traditional models widely employed in tumor research. In 1951, the initial human tumor cell line was isolated from an American cancer patient, which is widely known as HeLa cells [172]. CCLs span a variety of human tumors such as breast, central nervous system, colon, kidney, leukemia, lung, melanoma, ovary, and prostate [173]. Despite their diverse origins, these CCLs continue to play a crucial role in modern cancer research. PDTX is a valuable model which small fragments of tumors are surgically removed from cancer patients and then transplanted directly into immunodeficient mice [174]. In 1969, Ragaard and Povlsen successfully implanted human colonic tumor tissue into nude mice [175]. This achievement might be the first PDTX model. PDTXs are highly favored by researchers because they largely preserve the cellular and histopathological structures of the original tumors. In numerous preclinical cancer research studies, the presence of PDTXs is quite common. For example, in recently published studies in colorectal cancer, breast cancer [176], CC [177] and HCC [178]. Tumor organoids serve as a pivotal model system in the realm of precision medicine, owing to their unique capability to faithfully preserve the fundamental characteristics of the primary tumor [179]. They have emerged as a highly promising model for precision medicine, primarily due to their aptitude for predicting drug responses tailored to the unique profiles of individual patients. Tumor organoids can be effectively cultured from a diverse array of cancer types, encompassing brain, lung, liver, pancreatic, and breast cancer, among others [180]. The envisioned process involves procuring a patient’s tumor specimen, generating and sustaining organoids, exposing these patient-derived tumor organoids to a wide range of pharmaceutical agents, and subsequently administering the most optimal drug or drug combination. These diverse tumor models contribute to our understanding of cancer biology and aid in the development of potential treatments and they each have their respective advantages and disadvantages (Table 3).
The advantages and disadvantages of CCL, PDTX and PDO.
| Tumor models | Advantages | Disadvantages |
|---|---|---|
| CCL | Lower cost; Easy to maintain; High-throughput screening, Convenient manipulation |
Loss original tumor complexity; Lack tumor microenvironment; Lack genetic diversity |
| PDTX | Maintain tumor heterogeneity; Mimics in vivo microenvironment; More physiological environment; From human tumor |
Technically challenging; Expensive; Tumor restrictions; Long research time |
| PDO | Maintain tumor heterogeneity; Mimics in vivo microenvironment; Personalized medicine; Platform of drug discovery |
Longer culturing; Technically challenging; Expensive |
-
CCL, tumor cell line models; PDTX, patient-derived tumor xenograft models; PDO, patient-derived organoids.
Brain cancer
In a 2018 article published in the journal Nature Methods, Bian and their colleagues conducted a study to explore the use of genetically modified cerebral organoids as a reliable model for investigating the mechanisms involved in the formation of brain tumors [181]. The goal of this research was to enhance the utility of cerebral organoids by genetically modifying them to mimic the development of brain tumors. They introduced genetic mutations commonly associated with brain cancers, such as TP53 and PTEN alterations. This research is significant in the fields of cancer biology and neuro-oncology, providing a valuable platform for studying the molecular and cellular processes involved in brain tumor development. Glioblastomas are highly aggressive and deadly brain cancers known for their complexity and the presence of areas with low oxygen levels (hypoxic regions). In 2016, Hubert and their team conducted a study aimed at creating a 3D organoid culture system using human glioblastoma samples [182]. The main goal was to develop an in vitro model that accurately reproduces the hypoxic gradients and the diversity of cancer stem cells found in glioblastoma tumors in real-life conditions. The study successfully generated a 3D organoid culture system derived from human glioblastoma tissue. This organoid culture system shows promise as a valuable tool for studying glioblastomas and potentially other types of cancer.
Lung cancer
Lung cancer is a significant global cause of death. Lung cancer can be broadly classified into two main types: non-small cell lung cancer (NSCLC), and small cell lung cancer (SCLC). Within NSCLC, the most common subtype is lung adenocarcinoma (LUAD). It is a complex disease with diverse characteristics, both in terms of physical features and genetic makeup. This complexity makes it challenging to create animal models for research [183]. Researchers are increasingly using lung cancer organoids (LCOs) because they can faithfully replicate the structure and function of lung tissue found in the body.
LCOs are vital tools for researching the mechanisms of lung cancer. Zhang et al. demonstrated that the sustained expression and activation of KRAS, along with the loss of Lkb1, can transform normal lung organoids into malignant ones, promoting the transition to lung squamous cell carcinoma (LUSC) [184]. Subsequently, Dost and their colleagues used organoid systems derived from human iPSCs and mouse lung epithelial cells to simulate the development of LUAD. In both systems, the expression of KRAS in lung alveolar epithelial progenitor cells led to changes in the transcriptional program, resulting in the downregulation of genes associated with differentiation and maturation [185]. These two studies provided groundbreaking insights into the mechanisms of lung tumor formation. Kim et al. established various subtypes of LCOs, such as adenocarcinoma, squamous cell carcinoma, adenosquamous carcinoma, large cell carcinoma, and small cell carcinoma [186]. Li et al. [187] and Chen et al. [188] also successfully established biobanks of LCOs of different subtypes. These organoids exhibit a high degree of consistency with the original lung cancer tissue, offering new directions for personalized and precision therapies for lung cancer. Additionally, LCOs can be used for drug sensitivity testing. One study used 212 LCOs generated from 107 patients for drug sensitivity testing (LCO-DST). LCO-DST accurately predicted the clinical responses of some lung cancer patients to treatment [189]. Beyond drug sensitivity testing, LCOs can be applied in drug development by conducting high-throughput screening of numerous compounds to identify potential anti-cancer drugs [190].
Liver cancer
PLC is the second leading cause of cancer-related deaths worldwide. PLC encompasses various types, including HCC, intrahepatic cholangiocarcinoma (iCCA), and other less common tumors [191]. In cancer research, it is crucial to create accurate models to understand the disease better. Traditional 2D models have limitations, but 3D systems offer a more faithful representation of what happens in the body.
Hepatitis B virus (HBV) is a significant factor in the development of liver cancer. One study investigated liver cancer using PDO created from non-tumor cirrhotic liver tissues of liver transplant patients. Analysis of the gene expression patterns in these organoids revealed unusual early cancer-related characteristics. These findings have the potential to serve as new biological markers for understanding how HBV infection contributes to the development of liver cancer [192]. Naruse and colleagues established a novel in vitro model for inducing carcinogenesis through chemical treatments, using liver organoids. They used liver organoids derived from mouse tissues and exposed them to a chemical called diethylnitrosamine (DEN). This model helped explain the patterns of chemical carcinogenesis and how it can lead to liver cancer [193]. Sun et al. took a different approach by using reprogrammed human liver cells (hiHeps) with deactivated p53 and RB genes to create liver organoids. These organoids were then genetically modified to simulate the development of human liver cancer [194]. Li and their team highlighted the potential of tumor organoids in screening drugs for liver cancer treatment. They established 27 different liver cancer cell lines and tested them with 129 different cancer drugs. This research revealed significant variations in how tumors respond to these drugs, offering new insights into personalized treatment strategies for liver cancer [195]. These innovative approaches and organoid models are helping researchers better understand the complexities of liver cancer, providing new avenues for studying its development and potential treatments.
Pancreatic cancer
Pancreatic cancer is an extremely aggressive tumor with a very low 5-year survival rate. It ranks as the seventh most common cancer globally [196]. Pancreatic cancer organoids retain the physiological characteristics and functions of a patient’s original tumor cells. These organoids are valuable for personalized drug screening and drug sensitivity testing, offering promise for early diagnosis and personalized treatment of pancreatic cancer.
Researchers, such as those in the Clevers and Tuveson labs, have used surgically removed pancreatic cancer tissues to create pancreatic cancer organ models. They found that transplanting these organoids can mimic the full range of tumor development, offering insights into the entire cancer progression process [43]. One study discovered that the expression of the PDAC-related cancer gene GNASR201C induced cystic growth more effectively in pancreatic ducts compared to pancreatic acinar organoids. On the other hand, KRAS G12D was more effective at simulating cancer when expressed in acinar organoids as opposed to ductal organoids. The researchers developed a renewable source of pancreatic ductal and acinar organoids, providing a tool to study the lineage plasticity and human pancreatic cancer gene actions [197]. A team of researchers from Harvard Medical School, as published in Nature Medicine, employed 3D cell culture techniques to amplify and cultivate primary cancer cell organoids from pancreatic cancer patients’ tissues. These tumor organoids maintain the characteristics of the original tumor and preserve the specific physiological changes unique to the patients. These pancreatic tumor organoids can be used to simulate pancreatic cancer and conduct drug screening for precision treatments.
Breast cancer
Breast cancer is one of the most common malignancies affecting women globally and stands as the second leading cause of cancer-related deaths in women. The primary research models for breast cancer have traditionally been tumor CCL and PDTX. However, both of these models have their limitations. In 2018, Sachs and their team made a significant breakthrough by creating breast cancer organoids from pathological tissues of breast cancer patients. Furthermore, the team established a biobank comprising over 100 breast cancer organoid samples [198]. Building upon research conducted by Hans Clevers’ team, Mazzucchelli and others isolated breast cancer cells from surgical or biopsy samples and then cultivated them into organoids [199]. Li and colleagues successfully cultured organoids from large papillary breast cancer. These organoids continued to grow for over 6 months. These proliferating breast cancer organoids exhibited consistent pathology, hormone receptor expression, and the proliferation marker Ki-67 with post-surgery pathological results [200]. In another study, Nayak and their team cultured primary breast cancer cells from different patients in specific culture conditions, generating breast cancer organoids. They then exposed these organoids to chemotherapy drugs, doxorubicin, and mitoxantrone. The results highlighted significant differences in the response of organoids from different patients to these two chemotherapy drugs [201]. This underscores the capability of organoid models to replicate the tumor’s heterogeneity in vitro, offering a basis for personalized treatment approaches.
In summary, organoids derived from AdSCs, pluripotent stem cells, and tumor tissues have their own roles in human disease modeling. They can well simulate the characteristics of in vivo tissue biology, and capture some of the multicellular structure, anatomy, and even functional characteristics similar to real organs, providing an important platform for the study of disease modeling in different fields.
Conclusions and perspectives
Ever since Rheinwald and Green first reconstituted 3D tissue structures from cultured human stem cells [202], [203], [204], organoids are now defined as self-organized 3D structures grown from stem cells, composed of organ-specific cell types, and capable of reconstituting specific organ functions in vitro [205, 206]. After decades of development, organoids have been generated from various tissues, including the small intestine, prostate, brain, lung, and liver. Organoid technology has opened up new possibilities for scientific discovery in developmental biology, drug discovery, and personalized therapy [207], and multiple biobanks of organoids have been established for modeling human diseases and drug discovery, bringing the promise of personalized medicine closer to reality [36, 208] (Figure 2).
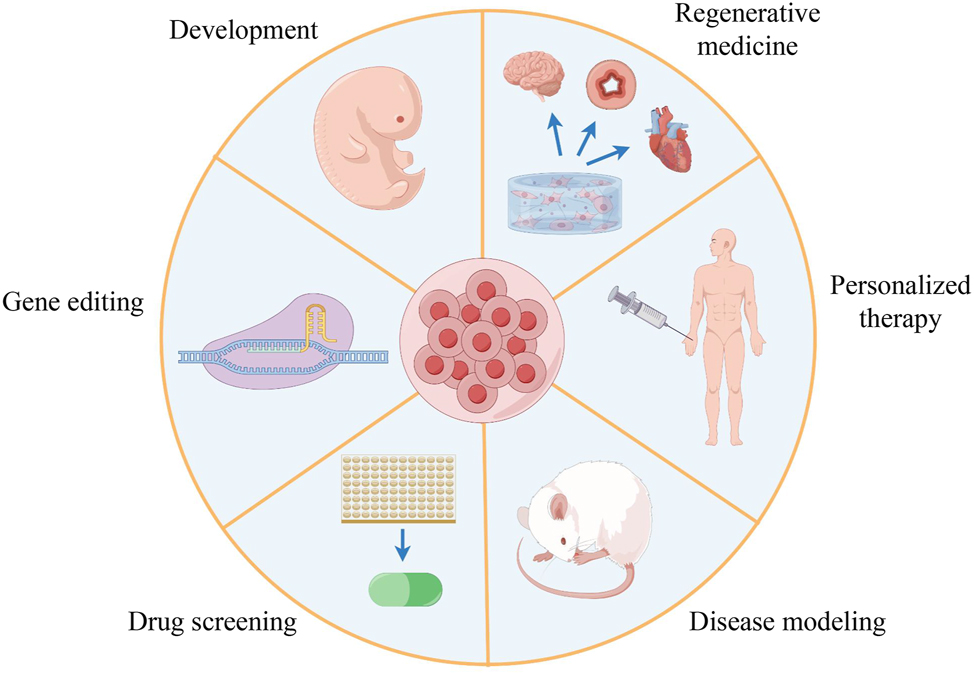
Multiple applications of organoid technology (by Figdraw). The schematic summarizes the different applications of organoids in many fields, including developmental biology, gene editing, drug screening, disease modeling, personalized therapy, and regenerative medicine.
The ethical issues of organoids cannot be ignored. The first ethical issue that is often discussed is that researchers should only carry out experiments with the consent of donors. If the research is carried out without the donor’s knowledge and completely separated from the donor’s personal data, it will weaken the clinical application of organoids. Informed consent from donors in organoid research means that they have control over the conduct of the experiment and the option to opt out. Interviews showed that the patients who participated in the research wanted to be informed about the experimental study of organoids, and wanted to know the related profits and drug pricing involved in the follow-up [209, 210]. Another issue is the ethical challenge of organoids commercialization. Patients usually donate their tissues for nothing, but in addition to the distrust of the research community, they also worry about the experimental data involved in subsequent organoids, profits, and whether the company will be overly profitable. These negative effects may be solved by providing benefits to the donor. It can be financial support or clinical special care such as experiencing new therapies based on organoid research [211]. The pertinent departments ought to enhance discourse regarding the ethics surrounding organoids and ensure patients are informed about organoids in an open and transparent manner. Furthermore, it is imperative to establish appropriate legislation to regulate the advancement of this technology in clinical practice. Improving ethical supervision, reducing public concerns, and improving the understanding of organoids to help organoids continue to develop on the premise of ethics [212].

Liver organoids and brain organoids obtained from iPSCs and ESCs (by Figdraw). Induced pluripotent stem cells (iPSCs) and embryonic stem cells (ESCs) respectively from the inner cell mass of blastocysts and somatic cell reprogramming through the cultivation of different numbers of days and cell factors to generate the corresponding organoids.

Overview of organoid types (by Figdraw). Starting cell types include embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), adult stem cells (ASCs), and tumor stem cells, which are aggregated into cell populations by self-organization. The cell population differentiates into three germ layers: endoderm, mesoderm, and ectoderm, which further specializes into various organoid types.
Organoids are typically derived from a single germ layer through step-wise differentiation, whereas an organ contains cells from multiple germ layers. Multi-tissue organoids [213, 214] or assembloids [215, 216] are established through the co-culture of cells derived from at least two germ layers (Figures 3 and 4). Significant progress has been made in understanding diseases such as cystic fibrosis, pancreatic cancer, colon cancer, Zika virus, and microcephaly with the use of brain organoids [217], [218], [219]. Despite these tremendous achievements and rapid development, organoids have a few limitations that hinder their broader application. Organoid cultures exhibit significant heterogeneity and variable complexity in cellular composition, can undergo poorly controlled morphogenesis, and often lack stromal, vascular, and immunological components [220]. Several achievements have been made in vascularized organoids [221], mainly through transplanting organoids into highly vascular tissues in immunodeficient mice [222] or culturing organoids on microfluidic devices that mimic vasculature [223, 224]. The ectopic expression of human ETS variant 2 (ETV2) has also contributed to the formation of a complex vascular-like network in human cortical organoids (hCOs) [78]. However, the formation of specific cell types and the simulation of functional systems are difficult to overcome for organoids, especially the nervous system. The problem is further complicated if the interaction between cells and tissues is also explored. In addition, there is also variability in organoids derived from patients of different ages and genetic backgrounds [20], and the differences in tumor-like tumors can also lead to heterogeneity in tumor results [36]. For the above vulnerabilities, CRISPR-Cas9 technology may be used to engineer organoids with an isogenetic background to reduce variability [225]. Every model has its own advantages and disadvantages, and organoids are not exceptional. Pay attention to the limitations when applying them.
Due to the random configuration of traditional 3D culture, it is difficult to precisely control organoids and their local environment [226, 227]. Organoids, in collaboration with engineers and organ-on-a-chip technology, provide a path to more controllable organoid systems [228], [229], [230], which have been used to create the required concentration gradients to regulate tissue patterning and development. Scaffolds made of biological materials can improve the abnormal tissue structure caused by the organoid self-organization [231]. A significant step has been taken in modeling the alveolar–capillary unit of the lung with a biomimetic microsystem that reconstitutes the critical functional alveolar-capillary interface of the human lung [232]. However, simulating the important physiological process of organoids in vivo vascularization and innervation is still one of the indispensable keys to understanding the mechanism of tumorigenesis, metastasis, or neurodegenerative disease [233]. In addition, the wide application of Matrigel cannot reflect the dynamic changes of biomechanics in organs. To solve this problem, some biological companies or laboratories have cooperated with engineering laboratories to develop synthetic substrates with both biophysical and biochemical properties [234]. Furthermore, the combination of organoids with other technologies, including CRISPR/Cas9 genome-editing technology [235], mass spectrometry, single-cell RNA sequencing, and cryo-electron microscopy (cryo-EM) [206], has further advanced our understanding of organoid assembly and organ development. In the future, the integration of organoids with other technologies may accelerate the development of organoids, simulating more complex human physiological and pathological situations and improving the accuracy of organoids to reproduce human physiological processes.
Funding source: National Key Research and Development Program of China
Award Identifier / Grant number: 2022YFA1103800
Award Identifier / Grant number: 2023YFE0210100
Award Identifier / Grant number: 2019YFA0904500
Funding source: Strategic Priority Research Program of the Chinese Academy of Sciences
Award Identifier / Grant number: XDB0480000
Funding source: National Natural Science Foundation Projects of China
Award Identifier / Grant number: 92157202
Award Identifier / Grant number: 32025010
Award Identifier / Grant number: 32241002
Award Identifier / Grant number: 92254301
Award Identifier / Grant number: 32261160376
Award Identifier / Grant number: 31970709
Award Identifier / Grant number: 32070729
Award Identifier / Grant number: 32100619
Award Identifier / Grant number: 32170747
Award Identifier / Grant number: 32322022
Award Identifier / Grant number: 32370782
Award Identifier / Grant number: 32371007
Award Identifier / Grant number: 32300608
Award Identifier / Grant number: 32300620
Funding source: Key Research Program, CAS
Award Identifier / Grant number: ZDBS-ZRKJZ-TLC003
Funding source: International Cooperation Program, CAS
Award Identifier / Grant number: 154144KYSB20200006
Award Identifier / Grant number: 2023A1515030231
Award Identifier / Grant number: 2022A1515110493
Award Identifier / Grant number: 2020B1212060052
Award Identifier / Grant number: 2021A1515012513
Award Identifier / Grant number: 2021B1515020096
Award Identifier / Grant number: 2022A1515012616
Award Identifier / Grant number: 2022A1515110951
Award Identifier / Grant number: 2023B1212120009
Funding source: CAS Project for Young Scientists in Basic Research
Award Identifier / Grant number: YSBR-075
Funding source: Guangdong Province Science and Technology Program
Award Identifier / Grant number: 2023B1111050005
Funding source: Guangzhou Science and Technology Program
Award Identifier / Grant number: 202102021037
Award Identifier / Grant number: 202102020827
Award Identifier / Grant number: 202102080066
Award Identifier / Grant number: 202206060002
Award Identifier / Grant number: 2023A04J0414
Funding source: CAS Youth Innovation Promotion Association
Award Identifier / Grant number: Y2021097
Award Identifier / Grant number: 2021355
Funding source: NSFC/RGC Joint Grant Scheme
Award Identifier / Grant number: N_CUHK 428/22
Funding source: the open research funds from the Sixth Affiliated Hospital of Guangzhou Medical University, Qingyuan People’s Hospital
Award Identifier / Grant number: 202301-203
Acknowledgment
Figures 2– 4 were created by Figdraw.com.
-
Research ethics: Not applicable.
-
Informed consent: Not applicable.
-
Author contributions: Xingguo Liu was responsible for the conception and design of the manuscript; Baodan Chen, Cijie Du, Mengfei Wang and Jingyi Guo wrote the manuscript; Baodan Chen made the figures and Cijie Du made the tables; Xingguo Liu supervised and edited the manuscript. All authors have read and approved the final manuscript.
-
Competing interests: The authors have no conflicts of interest to declare.
-
Research funding: This work was supported by the National Key Research and Development Program of China (2022YFA1103800, 2023YFE0210100, and 2019YFA0904500), the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB0480000), the National Natural Science Foundation Projects of China (92157202, 32025010, 32241002, 92254301, 32261160376, 31970709, 32070729, 32100619, 32170747, 32322022, 32370782, 32371007, 32300608, and 32300620), the Key Research Program, CAS (ZDBS-ZRKJZ-TLC003), the International Cooperation Program, CAS (154144KYSB20200006), the CAS Project for Young Scientists in Basic Research (YSBR-075), the Guangdong Province Science and Technology Program (2023B1111050005, 2023A1515030231, 2022A1515110493, 2020B1212060052, 2021A1515012513, 2021B1515020096, 2022A1515012616, 2022A1515110951, 2023B1212120009), the Guangzhou Science and Technology Program (202102021037, 202102020827, 202102080066, 202206060002 and 2023A04J0414), the CAS Youth Innovation Promotion Association (Y2021097 and 2021355), NSFC/RGC Joint Grant Scheme (N_CUHK 428/22), and the open research funds from the Sixth Affiliated Hospital of Guangzhou Medical University, Qingyuan People’s Hospital (202301-203).
-
Data availability: No new data were created or analysed in this article. Data sharing is not applicable to this article.
References
1. Till, JE, Mc, CE. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res 1961;14:213–22. https://doi.org/10.2307/3570892.Search in Google Scholar
2. Rheinwald, JG, Green, H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 1975;6:331–43. https://doi.org/10.1016/s0092-8674(75)80001-8.Search in Google Scholar PubMed
3. Evans, MJ, Kaufman, MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981;292:154–6. https://doi.org/10.1038/292154a0.Search in Google Scholar PubMed
4. Martin, GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 1981;78:7634–8. https://doi.org/10.1073/pnas.78.12.7634.Search in Google Scholar PubMed PubMed Central
5. Thomson, JA, Itskovitz-Eldor, J, Shapiro, SS, Waknitz, MA, Swiergiel, JJ, Marshall, VS, et al.. Embryonic stem cell lines derived from human blastocysts. Science 1998;282:1145–7. https://doi.org/10.1126/science.282.5391.1145.Search in Google Scholar PubMed
6. Zheng, P. Maintaining genomic stability in pluripotent stem cells. Genome Instab Dis 2020;1:5. https://doi.org/10.1007/s42764-019-00008-4.Search in Google Scholar
7. O’Connor, N, Mulliken, J, Banks-Schlegel, S, Kehinde, O, Green, H. Grafting of burns with cultured epithelium prepared from autologous epidermal cells. Lancet 1981;1:75–8. https://doi.org/10.1016/s0140-6736(81)90006-4.Search in Google Scholar
8. Wilson, HV. A new method by which sponges may be artificially reared. Science 1907;25:912–5. https://doi.org/10.1126/science.25.649.912.Search in Google Scholar PubMed
9. Weiss, P, Taylor, AC. Reconstitution of complete organs from single-cell suspensions of chick embryos in advanced stages of differentiation. Proc Natl Acad Sci U S A 1960;46:1177–85. https://doi.org/10.1073/pnas.46.9.1177.Search in Google Scholar PubMed PubMed Central
10. Li, ML, Aggeler, J, Farson, DA, Hatier, C, Hassell, J, Bissell, MJ. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc Natl Acad Sci U S A 1987;84:136–40. https://doi.org/10.1073/pnas.84.1.136.Search in Google Scholar PubMed PubMed Central
11. Sato, T, Vries, RG, Snippert, HJ, van de Wetering, M, Barker, N, Stange, DE, et al.. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262–5. https://doi.org/10.1038/nature07935.Search in Google Scholar PubMed
12. Eiraku, M, Watanabe, K, Matsuo-Takasaki, M, Kawada, M, Yonemura, S, Matsumura, M, et al.. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008;3:519–32. https://doi.org/10.1016/j.stem.2008.09.002.Search in Google Scholar PubMed
13. Takahashi, K, Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–76. https://doi.org/10.1016/j.cell.2006.07.024.Search in Google Scholar PubMed
14. Zhang, YS, Aleman, J, Shin, SR, Kilic, T, Kim, D, Mousavi Shaegh, SA, et al.. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A 2017;114:E2293–02. https://doi.org/10.1073/pnas.1612906114.Search in Google Scholar PubMed PubMed Central
15. Rossi, G, Manfrin, A, Lutolf, MP. Progress and potential in organoid research. Nat Rev Genet 2018;19:671–87. https://doi.org/10.1038/s41576-018-0051-9.Search in Google Scholar PubMed
16. Corro, C, Novellasdemunt, L, Li, VSW. A brief history of organoids. Am J Physiol Cell Physiol 2020;319:C151–65. https://doi.org/10.1152/ajpcell.00120.2020.Search in Google Scholar PubMed PubMed Central
17. Orkin, RW, Gehron, P, McGoodwin, EB, Martin, GR, Valentine, T, Swarm, R. A murine tumor producing a matrix of basement membrane. J Exp Med 1977;145:204–20. https://doi.org/10.1084/jem.145.1.204.Search in Google Scholar PubMed PubMed Central
18. McCracken, KW, Cata, EM, Crawford, CM, Sinagoga, KL, Schumacher, M, Rockich, BE, et al.. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014;516:400–4. https://doi.org/10.1038/nature13863.Search in Google Scholar PubMed PubMed Central
19. Simian, M, Hirai, Y, Navre, M, Werb, Z, Lochter, A, Bissell, MJ. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development 2001;128:3117–31. https://doi.org/10.1242/dev.128.16.3117.Search in Google Scholar PubMed PubMed Central
20. Lancaster, MA, Renner, M, Martin, CA, Wenzel, D, Bicknell, LS, Hurles, ME, et al.. Cerebral organoids model human brain development and microcephaly. Nature 2013;501:373–9. https://doi.org/10.1038/nature12517.Search in Google Scholar PubMed PubMed Central
21. Giobbe, GG, Crowley, C, Luni, C, Campinoti, S, Khedr, M, Kretzschmar, K, et al.. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat Commun 2019;10:5658. https://doi.org/10.1038/s41467-019-13605-4.Search in Google Scholar PubMed PubMed Central
22. Sorrentino, G, Rezakhani, S, Yildiz, E, Nuciforo, S, Heim, MH, Lutolf, MP, et al.. Mechano-modulatory synthetic niches for liver organoid derivation. Nat Commun 2020;11:3416. https://doi.org/10.1038/s41467-020-17161-0.Search in Google Scholar PubMed PubMed Central
23. Curvello, R, Kerr, G, Micati, DJ, Chan, WH, Raghuwanshi, VS, Rosenbluh, J, et al.. Engineered plant-based nanocellulose hydrogel for small intestinal organoid growth. Adv Sci 2020;8:2002135. https://doi.org/10.1002/advs.202002135.Search in Google Scholar PubMed PubMed Central
24. Sakaguchi, H, Kadoshima, T, Soen, M, Narii, N, Ishida, Y, Ohgushi, M, et al.. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun 2015;6:8896. https://doi.org/10.1038/ncomms9896.Search in Google Scholar PubMed PubMed Central
25. Eiraku, M, Takata, N, Ishibashi, H, Kawada, M, Sakakura, E, Okuda, S, et al.. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011;472:51–6. https://doi.org/10.1038/nature09941.Search in Google Scholar PubMed
26. Park, SE, Georgescu, A, Huh, D. Organoids-on-a-chip. Science 2019;364:960–5. https://doi.org/10.1126/science.aaw7894.Search in Google Scholar PubMed PubMed Central
27. Shirure, VS, Hughes, CCW, George, SC. Engineering vascularized organoid-on-a-chip models. Annu Rev Biomed Eng 2021;23:141–67. https://doi.org/10.1146/annurev-bioeng-090120-094330.Search in Google Scholar PubMed
28. Xian, C, Zhang, J, Zhao, S, Li, XG. Gut-on-a-chip for disease models. J Tissue Eng 2023;14:20417314221149882. https://doi.org/10.1177/20417314221149882.Search in Google Scholar PubMed PubMed Central
29. Cognetti, JS, Moen, MT, Brewer, MG, Bryan, MR, Tice, JD, McGrath, JL, et al.. A photonic biosensor-integrated tissue chip platform for real-time sensing of lung epithelial inflammatory markers. Lab Chip 2023;23:239–50. https://doi.org/10.1039/d2lc00864e.Search in Google Scholar PubMed PubMed Central
30. Tang, XY, Wu, S, Wang, D, Chu, C, Hong, Y, Tao, M, et al.. Human organoids in basic research and clinical applications. Signal Transduct Targeted Ther 2022;7:168. https://doi.org/10.1038/s41392-022-01024-9.Search in Google Scholar PubMed PubMed Central
31. Zhu, J, Ji, L, Chen, Y, Li, H, Huang, M, Dai, Z, et al.. Organoids and organs-on-chips: insights into predicting the efficacy of systemic treatment in colorectal cancer. Cell Death Discovery 2023;9:72. https://doi.org/10.1038/s41420-023-01354-9.Search in Google Scholar PubMed PubMed Central
32. Barker, N, van Es, JH, Kuipers, J, Kujala, P, van den Born, M, Cozijnsen, M, et al.. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007;449:1003–7. https://doi.org/10.1038/nature06196.Search in Google Scholar PubMed
33. Clevers, H. Modeling development and disease with organoids. Cell 2016;165:1586–97. https://doi.org/10.1016/j.cell.2016.05.082.Search in Google Scholar PubMed
34. Sato, T, Stange, DE, Ferrante, M, Vries, RG, Van Es, JH, Van den Brink, S, et al.. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011;141:1762–72. https://doi.org/10.1053/j.gastro.2011.07.050.Search in Google Scholar PubMed
35. Fujii, M, Matano, M, Toshimitsu, K, Takano, A, Mikami, Y, Nishikori, S, et al.. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 2018;23:787–93 e6. https://doi.org/10.1016/j.stem.2018.11.016.Search in Google Scholar PubMed
36. van de Wetering, M, Francies, HE, Francis, JM, Bounova, G, Iorio, F, Pronk, A, et al.. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161:933–45. https://doi.org/10.1016/j.cell.2015.03.053.Search in Google Scholar PubMed PubMed Central
37. Lamers, MM, Beumer, J, van der Vaart, J, Knoops, K, Puschhof, J, Breugem, TI, et al.. SARS-CoV-2 productively infects human gut enterocytes. Science 2020;369:50–4. https://doi.org/10.1126/science.abc1669.Search in Google Scholar PubMed PubMed Central
38. Marsee, A, Roos, FJM, Verstegen, MMA, Consortium, HPBO, Gehart, H, de Koning, E, et al.. Building consensus on definition and nomenclature of hepatic, pancreatic, and biliary organoids. Cell Stem Cell 2021;28:816–32. https://doi.org/10.1016/j.stem.2021.04.005.Search in Google Scholar PubMed
39. Huch, M, Dorrell, C, Boj, SF, van Es, JH, Li, VS, van de Wetering, M, et al.. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 2013;494:247–50. https://doi.org/10.1038/nature11826.Search in Google Scholar PubMed PubMed Central
40. Huch, M, Gehart, H, van Boxtel, R, Hamer, K, Blokzijl, F, Verstegen, MM, et al.. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160:299–312. https://doi.org/10.1016/j.cell.2014.11.050.Search in Google Scholar PubMed PubMed Central
41. Hu, H, Gehart, H, Artegiani, B, LO-I, C, Dekkers, F, Basak, O, et al.. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 2018;175:1591–606 e19. https://doi.org/10.1016/j.cell.2018.11.013.Search in Google Scholar PubMed
42. Peng, WC, Logan, CY, Fish, M, Anbarchian, T, Aguisanda, F, Alvarez-Varela, A, et al.. Inflammatory cytokine TNFalpha promotes the long-term expansion of primary hepatocytes in 3D Culture. Cell 2018;175:1607–19 e15. https://doi.org/10.1016/j.cell.2018.11.012.Search in Google Scholar PubMed PubMed Central
43. Boj, SF, Hwang, CI, Baker, LA, ChioII, Engle, DD, Corbo, V, et al.. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324–38. https://doi.org/10.1016/j.cell.2014.12.021.Search in Google Scholar PubMed PubMed Central
44. Broutier, L, Mastrogiovanni, G, Verstegen, MM, Francies, HE, Gavarro, LM, Bradshaw, CR, et al.. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med 2017;23:1424–35. https://doi.org/10.1038/nm.4438.Search in Google Scholar PubMed PubMed Central
45. Adams, TS, Schupp, JC, Poli, S, Ayaub, EA, Neumark, N, Ahangari, F, et al.. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv 2020;6:eaba1983. https://doi.org/10.1126/sciadv.aba1983.Search in Google Scholar PubMed PubMed Central
46. Barkauskas, CE, Chung, MI, Fioret, B, Gao, X, Katsura, H, Hogan, BL. Lung organoids: current uses and future promise. Development 2017;144:986–97. https://doi.org/10.1242/dev.140103.Search in Google Scholar PubMed PubMed Central
47. Lu, T, Cao, Y, Zhao, P, Shen, S, Xi, Y. Organoid: a powerful tool to study lung regeneration and disease. Cell Regen 2021;10:21. https://doi.org/10.1186/s13619-021-00082-8.Search in Google Scholar PubMed PubMed Central
48. Nikolic, MZ, Sun, D, Rawlins, EL. Human lung development: recent progress and new challenges. Development 2018;145:dev163485. https://doi.org/10.1242/dev.163485.Search in Google Scholar PubMed PubMed Central
49. Rock, JR, Gao, X, Xue, Y, Randell, SH, Kong, YY, Hogan, BL. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 2011;8:639–48. https://doi.org/10.1016/j.stem.2011.04.003.Search in Google Scholar PubMed PubMed Central
50. Rock, JR, Onaitis, MW, Rawlins, EL, Lu, Y, Clark, CP, Xue, Y, et al.. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A 2009;106:12771–5. https://doi.org/10.1073/pnas.0906850106.Search in Google Scholar PubMed PubMed Central
51. Danahay, H, Pessotti, AD, Coote, J, Montgomery, BE, Xia, D, Wilson, A, et al.. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep 2015;10:239–52. https://doi.org/10.1016/j.celrep.2014.12.017.Search in Google Scholar PubMed
52. Hild, M, Jaffe, AB. Production of 3-D airway organoids from primary human airway basal cells and their use in high-throughput screening. Curr Protoc Stem Cell Biol 2016;37:IE 9 1–15. https://doi.org/10.1002/cpsc.1.Search in Google Scholar PubMed
53. McQualter, JL, Yuen, K, Williams, B, Bertoncello, I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci U S A 2010;107:1414–9. https://doi.org/10.1073/pnas.0909207107.Search in Google Scholar PubMed PubMed Central
54. Barkauskas, CE, Cronce, MJ, Rackley, CR, Bowie, EJ, Keene, DR, Stripp, BR, et al.. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 2013;123:3025–36. https://doi.org/10.1172/jci68782.Search in Google Scholar
55. Gonzalez, RF, Allen, L, Gonzales, L, Ballard, PL, Dobbs, LG. HTII-280, a biomarker specific to the apical plasma membrane of human lung alveolar type II cells. J Histochem Cytochem 2010;58:891–901. https://doi.org/10.1369/jhc.2010.956433.Search in Google Scholar PubMed PubMed Central
56. Mulay, A, Konda, B, Garcia, GJr., Yao, C, Beil, S, Villalba, JM, et al.. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep 2021;35:109055. https://doi.org/10.1016/j.celrep.2021.109055.Search in Google Scholar PubMed PubMed Central
57. Youk, J, Kim, T, Evans, KV, Jeong, YI, Hur, Y, Hong, SP, et al.. Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell 2020;27:905–19 e10. https://doi.org/10.1016/j.stem.2020.10.004.Search in Google Scholar PubMed PubMed Central
58. Zhao, D, Lei, W, Hu, S. Cardiac organoid – a promising perspective of preclinical model. Stem Cell Res Ther 2021;12:272. https://doi.org/10.1186/s13287-021-02340-7.Search in Google Scholar PubMed PubMed Central
59. Hofbauer, P, Jahnel, SM, Papai, N, Giesshammer, M, Deyett, A, Schmidt, C, et al.. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 2021;184:3299–317 e22. https://doi.org/10.1016/j.cell.2021.04.034.Search in Google Scholar PubMed
60. Lee, J, Sutani, A, Kaneko, R, Takeuchi, J, Sasano, T, Kohda, T, et al.. In vitro generation of functional murine heart organoids via FGF4 and extracellular matrix. Nat Commun 2020;11:4283. https://doi.org/10.1038/s41467-020-18031-5.Search in Google Scholar PubMed PubMed Central
61. Richards, DJ, Li, Y, Kerr, CM, Yao, J, Beeson, GC, Coyle, RC, et al.. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat Biomed Eng 2020;4:446–62. https://doi.org/10.1038/s41551-020-0539-4.Search in Google Scholar PubMed PubMed Central
62. Voges, HK, Mills, RJ, Elliott, DA, Parton, RG, Porrello, ER, Hudson, JE. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development 2017;144:1118–27. https://doi.org/10.1242/dev.143966.Search in Google Scholar PubMed
63. Goldfracht, I, Protze, S, Shiti, A, Setter, N, Gruber, A, Shaheen, N, et al.. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat Commun 2020;11:75. https://doi.org/10.1038/s41467-019-13868-x.Search in Google Scholar PubMed PubMed Central
64. Long, C, Li, H, Tiburcy, M, Rodriguez-Caycedo, C, Kyrychenko, V, Zhou, H, et al.. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv 2018;4:eaap9004. https://doi.org/10.1126/sciadv.aap9004.Search in Google Scholar PubMed PubMed Central
65. Sun, T, Hevner, RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci 2014;15:217–32. https://doi.org/10.1038/nrn3707.Search in Google Scholar PubMed PubMed Central
66. Shi, Y, Wu, Q, Wang, X. Modeling brain development and diseases with human cerebral organoids. Curr Opin Neurobiol 2021;66:103–15. https://doi.org/10.1016/j.conb.2020.09.006.Search in Google Scholar PubMed
67. Muguruma, K, Nishiyama, A, Kawakami, H, Hashimoto, K, Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep 2015;10:537–50. https://doi.org/10.1016/j.celrep.2014.12.051.Search in Google Scholar PubMed
68. Miura, Y, Li, MY, Birey, F, Ikeda, K, Revah, O, Thete, MV, et al.. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat Biotechnol 2020;38:1421–30. https://doi.org/10.1038/s41587-020-00763-w.Search in Google Scholar PubMed PubMed Central
69. Huang, WK, Wong, SZH, Pather, SR, Nguyen, PTT, Zhang, F, Zhang, DY, et al.. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell 2021;28:1657–70 e10. https://doi.org/10.1016/j.stem.2021.04.006.Search in Google Scholar PubMed PubMed Central
70. Xiang, Y, Tanaka, Y, Cakir, B, Patterson, B, Kim, KY, Sun, P, et al.. hESC-Derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell 2019;24:487–97 e7. https://doi.org/10.1016/j.stem.2018.12.015.Search in Google Scholar PubMed PubMed Central
71. Silva, TP, Fernandes, TG, Nogueira, DES, Rodrigues, CAV, Bekman, EP, Hashimura, Y, et al.. Scalable generation of mature cerebellar organoids from human pluripotent stem cells and characterization by immunostaining. J Vis Exp 2020;e61143. https://doi.org/10.3791/61143.Search in Google Scholar PubMed
72. Jo, J, Xiao, Y, Sun, AX, Cukuroglu, E, Tran, HD, Goke, J, et al.. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 2016;19:248–57. https://doi.org/10.1016/j.stem.2016.07.005.Search in Google Scholar PubMed PubMed Central
73. Kadoshima, T, Sakaguchi, H, Nakano, T, Soen, M, Ando, S, Eiraku, M, et al.. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A 2013;110:20284–9. https://doi.org/10.1073/pnas.1315710110.Search in Google Scholar PubMed PubMed Central
74. Amin, ND, Pasca, SP. Building models of brain disorders with three-dimensional organoids. Neuron 2018;100:389–405. https://doi.org/10.1016/j.neuron.2018.10.007.Search in Google Scholar PubMed
75. Klaus, J, Kanton, S, Kyrousi, C, Ayo-Martin, AC, Di Giaimo, R, Riesenberg, S, et al.. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat Med 2019;25:561–8. https://doi.org/10.1038/s41591-019-0371-0.Search in Google Scholar PubMed
76. He, Z, Maynard, A, Jain, A, Gerber, T, Petri, R, Lin, HC, et al.. Lineage recording in human cerebral organoids. Nat Methods 2022;19:90–9. https://doi.org/10.1038/s41592-021-01344-8.Search in Google Scholar PubMed PubMed Central
77. Bagley, JA, Reumann, D, Bian, S, Levi-Strauss, J, Knoblich, JA. Fused cerebral organoids model interactions between brain regions. Nat Methods 2017;14:743–51. https://doi.org/10.1038/nmeth.4304.Search in Google Scholar PubMed PubMed Central
78. Cakir, B, Xiang, Y, Tanaka, Y, Kural, MH, Parent, M, Kang, YJ, et al.. Engineering of human brain organoids with a functional vascular-like system. Nat Methods 2019;16:1169–75. https://doi.org/10.1038/s41592-019-0586-5.Search in Google Scholar PubMed PubMed Central
79. Dobyns, WB, Stratton, RF, Parke, JT, Greenberg, F, Nussbaum, RL, Ledbetter, DH. Miller-Dieker syndrome: lissencephaly and monosomy 17p. J Pediatr 1983;102:552–8. https://doi.org/10.1016/s0022-3476(83)80183-8.Search in Google Scholar PubMed
80. Nagamani, SC, Zhang, F, Shchelochkov, OA, Bi, W, Ou, Z, Scaglia, F, et al.. Microdeletions including YWHAE in the Miller-Dieker syndrome region on chromosome 17p13.3 result in facial dysmorphisms, growth restriction, and cognitive impairment. J Med Genet 2009;46:825–33. https://doi.org/10.1136/jmg.2009.067637.Search in Google Scholar PubMed
81. Cardoso, C, Leventer, RJ, Ward, HL, Toyo-Oka, K, Chung, J, Gross, A, et al.. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet 2003;72:918–30. https://doi.org/10.1086/374320.Search in Google Scholar PubMed PubMed Central
82. Bershteyn, M, Nowakowski, TJ, Pollen, AA, Di Lullo, E, Nene, A, Wynshaw-Boris, A, et al.. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 2017;20:435–49 e4. https://doi.org/10.1016/j.stem.2016.12.007.Search in Google Scholar PubMed PubMed Central
83. Iefremova, V, Manikakis, G, Krefft, O, Jabali, A, Weynans, K, Wilkens, R, et al.. An organoid-based model of cortical development identifies non-cell-autonomous defects in Wnt signaling contributing to Miller-Dieker syndrome. Cell Rep 2017;19:50–9. https://doi.org/10.1016/j.celrep.2017.03.047.Search in Google Scholar PubMed
84. Borrell, V, Gotz, M. Role of radial glial cells in cerebral cortex folding. Curr Opin Neurobiol 2014;27:39–46. https://doi.org/10.1016/j.conb.2014.02.007.Search in Google Scholar PubMed
85. Nowakowski, TJ, Pollen, AA, Sandoval-Espinosa, C, Kriegstein, AR. Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron 2016;91:1219–27. https://doi.org/10.1016/j.neuron.2016.09.005.Search in Google Scholar PubMed PubMed Central
86. Nawathe, A, Doherty, J, Pandya, P. Fetal microcephaly. BMJ 2018;361:k2232. https://doi.org/10.1136/bmj.k2232.Search in Google Scholar PubMed
87. Bond, J, Roberts, E, Springell, K, Lizarraga, SB, Scott, S, Higgins, J, et al.. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet 2005;37:353–5. https://doi.org/10.1038/ng1539.Search in Google Scholar PubMed
88. Bond, J, Roberts, E, Mochida, GH, Hampshire, DJ, Scott, S, Askham, JM, et al.. ASPM is a major determinant of cerebral cortical size. Nat Genet 2002;32:316–20. https://doi.org/10.1038/ng995.Search in Google Scholar PubMed
89. Jayaraman, D, Bae, BI, Walsh, CA. The genetics of primary microcephaly. Annu Rev Genom Hum Genet 2018;19:177–200. https://doi.org/10.1146/annurev-genom-083117-021441.Search in Google Scholar PubMed
90. Barrera, JA, Kao, LR, Hammer, RE, Seemann, J, Fuchs, JL, Megraw, TL. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell 2010;18:913–26. https://doi.org/10.1016/j.devcel.2010.05.017.Search in Google Scholar PubMed PubMed Central
91. Li, R, Sun, L, Fang, A, Li, P, Wu, Q, Wang, X. Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell 2017;8:823–33. https://doi.org/10.1007/s13238-017-0479-2.Search in Google Scholar PubMed PubMed Central
92. Wang, L, Li, Z, Sievert, D, Smith, DEC, Mendes, MI, Chen, DY, et al.. Loss of NARS1 impairs progenitor proliferation in cortical brain organoids and leads to microcephaly. Nat Commun 2020;11:4038. https://doi.org/10.1038/s41467-020-17454-4.Search in Google Scholar PubMed PubMed Central
93. Esk, C, Lindenhofer, D, Haendeler, S, Wester, RA, Pflug, F, Schroeder, B, et al.. A human tissue screen identifies a regulator of ER secretion as a brain-size determinant. Science 2020;370:935–41. https://doi.org/10.1126/science.abb5390.Search in Google Scholar PubMed
94. Garcez, PP, Loiola, EC, Madeiro da Costa, R, Higa, LM, Trindade, P, Delvecchio, R, et al.. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016;352:816–8. https://doi.org/10.1126/science.aaf6116.Search in Google Scholar PubMed
95. Yoon, KJ, Song, G, Qian, X, Pan, J, Xu, D, Rho, HS, et al.. Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell 2017;21:349–58 e6. https://doi.org/10.1016/j.stem.2017.07.014.Search in Google Scholar PubMed PubMed Central
96. Liang, Q, Luo, Z, Zeng, J, Chen, W, Foo, SS, Lee, SA, et al.. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 2016;19:663–71. https://doi.org/10.1016/j.stem.2016.07.019.Search in Google Scholar PubMed PubMed Central
97. Zhou, T, Tan, L, Cederquist, GY, Fan, Y, Hartley, BJ, Mukherjee, S, et al.. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 2017;21:274–83 e5. https://doi.org/10.1016/j.stem.2017.06.017.Search in Google Scholar PubMed PubMed Central
98. Xu, M, Lee, EM, Wen, Z, Cheng, Y, Huang, WK, Qian, X, et al.. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 2016;22:1101–7. https://doi.org/10.1038/nm.4184.Search in Google Scholar PubMed PubMed Central
99. Hallmayer, J, Cleveland, S, Torres, A, Phillips, J, Cohen, B, Torigoe, T, et al.. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatr 2011;68:1095–102. https://doi.org/10.1001/archgenpsychiatry.2011.76.Search in Google Scholar PubMed PubMed Central
100. Mariani, J, Coppola, G, Zhang, P, Abyzov, A, Provini, L, Tomasini, L, et al.. FOXG1-Dependent dysregulation of GABA/Glutamate neuron differentiation in autism spectrum disorders. Cell 2015;162:375–90. https://doi.org/10.1016/j.cell.2015.06.034.Search in Google Scholar PubMed PubMed Central
101. Wang, P, Mokhtari, R, Pedrosa, E, Kirschenbaum, M, Bayrak, C, Zheng, D, et al.. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol Autism 2017;8:11. https://doi.org/10.1186/s13229-017-0124-1.Search in Google Scholar PubMed PubMed Central
102. Paulsen, B, Velasco, S, Kedaigle, AJ, Pigoni, M, Quadrato, G, Deo, AJ, et al.. Autism genes converge on asynchronous development of shared neuron classes. Nature 2022;602:268–73. https://doi.org/10.1038/s41586-021-04358-6.Search in Google Scholar PubMed PubMed Central
103. Wilson, DM3rd, Cookson, MR, Van Den Bosch, L, Zetterberg, H, Holtzman, DM, Dewachter, I. Hallmarks of neurodegenerative diseases. Cell 2023;186:693–714. https://doi.org/10.1016/j.cell.2022.12.032.Search in Google Scholar PubMed
104. Dugger, BN, Dickson, DW. Pathology of neurodegenerative diseases. Cold Spring Harbor Perspect Biol 2017;9:a028035. https://doi.org/10.1101/cshperspect.a028035.Search in Google Scholar PubMed PubMed Central
105. Arvanitakis, Z, Shah, RC, Bennett, DA. Diagnosis and management of dementia: review. JAMA 2019;322:1589–99. https://doi.org/10.1001/jama.2019.4782.Search in Google Scholar PubMed PubMed Central
106. Hanseeuw, BJ, Betensky, RA, Jacobs, HIL, Schultz, AP, Sepulcre, J, Becker, JA, et al.. Association of amyloid and Tau with cognition in preclinical Alzheimer disease: a longitudinal study. JAMA Neurol 2019;76:915–24. https://doi.org/10.1001/jamaneurol.2019.1424.Search in Google Scholar PubMed PubMed Central
107. Zhao, H, Wang, S, Li, X. DNA damage accumulation in aging brain and its links to Alzheimer’s disease progression. Genome Instab Dis 2022;3:7. https://doi.org/10.1007/s42764-022-00069-y.Search in Google Scholar
108. Raja, WK, Mungenast, AE, Lin, YT, Ko, T, Abdurrob, F, Seo, J, et al.. Self-Organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS One 2016;11:e0161969. https://doi.org/10.1371/journal.pone.0161969.Search in Google Scholar PubMed PubMed Central
109. Kuehner, JN, Chen, J, Bruggeman, EC, Wang, F, Li, Y, Xu, C, et al.. 5-hydroxymethylcytosine is dynamically regulated during forebrain organoid development and aberrantly altered in Alzheimer’s disease. Cell Rep 2021;35:109042. https://doi.org/10.1016/j.celrep.2021.109042.Search in Google Scholar PubMed PubMed Central
110. Shimada, H, Sato, Y, Sasaki, T, Shimozawa, A, Imaizumi, K, Shindo, T, et al.. A next-generation iPSC-derived forebrain organoid model of tauopathy with tau fibrils by AAV-mediated gene transfer. Cells Rep Methods 2022;2:100289. https://doi.org/10.1016/j.crmeth.2022.100289.Search in Google Scholar PubMed PubMed Central
111. Yin, J, VanDongen, AM. Enhanced neuronal activity and asynchronous calcium transients revealed in a 3D organoid model of Alzheimer’s disease. ACS Biomater Sci Eng 2021;7:254–64. https://doi.org/10.1021/acsbiomaterials.0c01583.Search in Google Scholar PubMed
112. Park, J, Wetzel, I, Marriott, I, Dreau, D, D’Avanzo, C, Kim, DY, et al.. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci 2018;21:941–51. https://doi.org/10.1038/s41593-018-0175-4.Search in Google Scholar PubMed PubMed Central
113. Huang, S, Zhang, Z, Cao, J, Yu, Y, Pei, G. Chimeric cerebral organoids reveal the essentials of neuronal and astrocytic APOE4 for Alzheimer’s tau pathology. Signal Transduct Targeted Ther 2022;7:176. https://doi.org/10.1038/s41392-022-01006-x.Search in Google Scholar PubMed PubMed Central
114. Perez, MJ, Ivanyuk, D, Panagiotakopoulou, V, Di Napoli, G, Kalb, S, Brunetti, D, et al.. Loss of function of the mitochondrial peptidase PITRM1 induces proteotoxic stress and Alzheimer’s disease-like pathology in human cerebral organoids. Mol Psychiatr 2021;26:5733–50. https://doi.org/10.1038/s41380-020-0807-4.Search in Google Scholar PubMed PubMed Central
115. Zhao, J, Fu, Y, Yamazaki, Y, Ren, Y, Davis, MD, Liu, CC, et al.. APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer’s disease patient iPSC-derived cerebral organoids. Nat Commun 2020;11:5540. https://doi.org/10.1038/s41467-020-19264-0.Search in Google Scholar PubMed PubMed Central
116. Choi, H, Kim, HJ, Yang, J, Chae, S, Lee, W, Chung, S, et al.. Acetylation changes tau interactome to degrade tau in Alzheimer’s disease animal and organoid models. Aging Cell 2020;19:e13081. https://doi.org/10.1111/acel.13081.Search in Google Scholar PubMed PubMed Central
117. Park, JC, Jang, SY, Lee, D, Lee, J, Kang, U, Chang, H, et al.. A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids. Nat Commun 2021;12:280. https://doi.org/10.1038/s41467-020-20440-5.Search in Google Scholar PubMed PubMed Central
118. Reddy, KS. Global burden of disease study 2015 provides GPS for global health 2030. Lancet 2016;388:1448–9. https://doi.org/10.1016/s0140-6736(16)31743-3.Search in Google Scholar
119. Tolosa, E, Garrido, A, Scholz, SW, Poewe, W. Challenges in the diagnosis of Parkinson’s disease. Lancet Neurol 2021;20:385–97. https://doi.org/10.1016/s1474-4422(21)00030-2.Search in Google Scholar PubMed PubMed Central
120. Spillantini, MG, Schmidt, ML, Lee, VM, Trojanowski, JQ, Jakes, R, Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997;388:839–40. https://doi.org/10.1038/42166.Search in Google Scholar PubMed
121. Mohamed, NV, Sirois, J, Ramamurthy, J, Mathur, M, Lepine, P, Deneault, E, et al.. Midbrain organoids with an SNCA gene triplication model key features of synucleinopathy. Brain Commun 2021;3:fcab223. https://doi.org/10.1093/braincomms/fcab223.Search in Google Scholar PubMed PubMed Central
122. Smits, LM, Reinhardt, L, Reinhardt, P, Glatza, M, Monzel, AS, Stanslowsky, N, et al.. Modeling Parkinson’s disease in midbrain-like organoids. npj Parkinson’s Dis 2019;5:5. https://doi.org/10.1038/s41531-019-0078-4.Search in Google Scholar PubMed PubMed Central
123. Wulansari, N, Darsono, WHW, Woo, HJ, Chang, MY, Kim, J, Bae, EJ, et al.. Neurodevelopmental defects and neurodegenerative phenotypes in human brain organoids carrying Parkinson’s disease-linked DNAJC6 mutations. Sci Adv 2021;7:eabb1540. https://doi.org/10.1126/sciadv.abb1540.Search in Google Scholar PubMed PubMed Central
124. Jo, J, Yang, L, Tran, HD, Yu, W, Sun, AX, Chang, YY, et al.. Lewy Body-like Inclusions in human midbrain organoids carrying glucocerebrosidase and alpha-Synuclein mutations. Ann Neurol 2021;90:490–505. https://doi.org/10.1002/ana.26166.Search in Google Scholar PubMed PubMed Central
125. Jarazo, J, Barmpa, K, Modamio, J, Saraiva, C, Sabate-Soler, S, Rosety, I, et al.. Parkinson’s disease phenotypes in patient neuronal cultures and brain organoids improved by 2-Hydroxypropyl-beta-cyclodextrin treatment. Mov Disord 2022;37:80–94. https://doi.org/10.1002/mds.28810.Search in Google Scholar PubMed PubMed Central
126. Goutman, SA, Hardiman, O, Al-Chalabi, A, Chio, A, Savelieff, MG, Kiernan, MC, et al.. Recent advances in the diagnosis and prognosis of amyotrophic lateral sclerosis. Lancet Neurol 2022;21:480–93. https://doi.org/10.1016/s1474-4422(21)00465-8.Search in Google Scholar PubMed PubMed Central
127. van Es, MA, Hardiman, O, Chio, A, Al-Chalabi, A, Pasterkamp, RJ, Veldink, JH, et al.. Amyotrophic lateral sclerosis. Lancet 2017;390:2084–98. https://doi.org/10.1016/S0140-6736(17)31287-4.Search in Google Scholar PubMed
128. Ling, SC, Polymenidou, M, Cleveland, DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 2013;79:416–38. https://doi.org/10.1016/j.neuron.2013.07.033.Search in Google Scholar PubMed PubMed Central
129. Wang, H, Guan, L, Deng, M. Recent progress of the genetics of amyotrophic lateral sclerosis and challenges of gene therapy. Front Neurosci 2023;17:1170996. https://doi.org/10.3389/fnins.2023.1170996.Search in Google Scholar PubMed PubMed Central
130. Szebenyi, K, Wenger, LMD, Sun, Y, Dunn, AWE, Limegrover, CA, Gibbons, GM, et al.. Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nat Neurosci 2021;24:1542–54. https://doi.org/10.1038/s41593-021-00923-4.Search in Google Scholar PubMed PubMed Central
131. Hong, Y, Dong, X, Chang, L, Xie, C, Chang, M, Aguilar, JS, et al.. Microglia-containing cerebral organoids derived from induced pluripotent stem cells for the study of neurological diseases. iScience 2023;26:106267. https://doi.org/10.1016/j.isci.2023.106267.Search in Google Scholar PubMed PubMed Central
132. Pereira, JD, DuBreuil, DM, Devlin, AC, Held, A, Sapir, Y, Berezovski, E, et al.. Human sensorimotor organoids derived from healthy and amyotrophic lateral sclerosis stem cells form neuromuscular junctions. Nat Commun 2021;12:4744. https://doi.org/10.1038/s41467-021-24776-4.Search in Google Scholar PubMed PubMed Central
133. Duncan, AW, Dorrell, C, Grompe, M. Stem cells and liver regeneration. Gastroenterology 2009;137:466–81. https://doi.org/10.1053/j.gastro.2009.05.044.Search in Google Scholar PubMed PubMed Central
134. Asrani, SK, Devarbhavi, H, Eaton, J, Kamath, PS. Burden of liver diseases in the world. J Hepatol 2019;70:151–71. https://doi.org/10.1016/j.jhep.2018.09.014.Search in Google Scholar PubMed
135. Heslop, JA, Rowe, C, Walsh, J, Sison-Young, R, Jenkins, R, Kamalian, L, et al.. Mechanistic evaluation of primary human hepatocyte culture using global proteomic analysis reveals a selective dedifferentiation profile. Arch Toxicol 2017;91:439–52. https://doi.org/10.1007/s00204-016-1694-y.Search in Google Scholar PubMed PubMed Central
136. Gural, N, Mancio-Silva, L, He, J, Bhatia, SN. Engineered livers for infectious diseases. Cell Mol Gastroenterol Hepatol 2018;5:131–44. https://doi.org/10.1016/j.jcmgh.2017.11.005.Search in Google Scholar PubMed PubMed Central
137. Prior, N, Inacio, P, Huch, M. Liver organoids: from basic research to therapeutic applications. Gut 2019;68:2228–37. https://doi.org/10.1136/gutjnl-2019-319256.Search in Google Scholar PubMed PubMed Central
138. Takebe, T, Sekine, K, Enomura, M, Koike, H, Kimura, M, Ogaeri, T, et al.. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013;499:481–4. https://doi.org/10.1038/nature12271.Search in Google Scholar PubMed
139. Nuciforo, S, Heim, MH. Organoids to model liver disease. JHEP Rep 2021;3:100198. https://doi.org/10.1016/j.jhepr.2020.100198.Search in Google Scholar PubMed PubMed Central
140. Rowe, RG, Daley, GQ. Induced pluripotent stem cells in disease modelling and drug discovery. Nat Rev Genet 2019;20:377–88. https://doi.org/10.1038/s41576-019-0100-z.Search in Google Scholar PubMed PubMed Central
141. Ouchi, R, Togo, S, Kimura, M, Shinozawa, T, Koido, M, Koike, H, et al.. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metabol 2019;30:374–84 e6. https://doi.org/10.1016/j.cmet.2019.05.007.Search in Google Scholar PubMed PubMed Central
142. Fiorotto, R, Amenduni, M, Mariotti, V, Fabris, L, Spirli, C, Strazzabosco, M. Liver diseases in the dish: iPSC and organoids as a new approach to modeling liver diseases. Biochim Biophys Acta Mol Basis Dis 2019;1865:920–8. https://doi.org/10.1016/j.bbadis.2018.08.038.Search in Google Scholar PubMed PubMed Central
143. Drucker, DJ, Jin, T, Asa, SL, Young, TA, Brubaker, PL. Activation of proglucagon gene transcription by protein kinase-A in a novel mouse enteroendocrine cell line. Mol Endocrinol 1994;8:1646–55. https://doi.org/10.1210/mend.8.12.7535893.Search in Google Scholar PubMed
144. Evers, BM, Townsend, CMJr., Upp, JR, Allen, E, Hurlbut, SC, Kim, SW, et al.. Establishment and characterization of a human carcinoid in nude mice and effect of various agents on tumor growth. Gastroenterology 1991;101:303–11. https://doi.org/10.1016/0016-5085(91)90004-5.Search in Google Scholar PubMed
145. Crespo, M, Vilar, E, Tsai, SY, Chang, K, Amin, S, Srinivasan, T, et al.. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat Med 2017;23:878–84. https://doi.org/10.1038/nm.4355.Search in Google Scholar PubMed PubMed Central
146. Faller, WJ, Jackson, TJ, Knight, JR, Ridgway, RA, Jamieson, T, Karim, SA, et al.. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 2015;517:497–500. https://doi.org/10.1038/nature13896.Search in Google Scholar PubMed PubMed Central
147. Huang, SM, Mishina, YM, Liu, S, Cheung, A, Stegmeier, F, Michaud, GA, et al.. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009;461:614–20. https://doi.org/10.1038/nature08356.Search in Google Scholar PubMed
148. Zhang, H, Li, HB, Lyu, JR, Lei, XM, Li, W, Wu, G, et al.. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int J Infect Dis 2020;96:19–24. https://doi.org/10.1016/j.ijid.2020.04.027.Search in Google Scholar PubMed PubMed Central
149. Kruger, J, Gross, R, Conzelmann, C, Muller, JA, Koepke, L, Sparrer, KMJ, et al.. Drug inhibition of SARS-CoV-2 replication in human pluripotent stem cell-derived intestinal organoids. Cell Mol Gastroenterol Hepatol 2021;11:935–48. https://doi.org/10.1016/j.jcmgh.2020.11.003.Search in Google Scholar PubMed PubMed Central
150. Csobonyeiova, M, Klein, M, Kuniakova, M, Varga, I, Danisovic, L. Induced pluripotent stem cell-derived organoids: their implication in COVID-19 modeling. Int J Mol Sci 2023;24:3459. https://doi.org/10.3390/ijms24043459.Search in Google Scholar PubMed PubMed Central
151. Li, M, Gong, J, Gao, L, Zou, T, Kang, J, Xu, H. Advanced human developmental toxicity and teratogenicity assessment using human organoid models. Ecotoxicol Environ Saf 2022;235:113429. https://doi.org/10.1016/j.ecoenv.2022.113429.Search in Google Scholar PubMed
152. Qu, M, Xiong, L, Lyu, Y, Zhang, X, Shen, J, Guan, J, et al.. Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Res 2021;31:259–71. https://doi.org/10.1038/s41422-020-00453-x.Search in Google Scholar PubMed PubMed Central
153. Holloway, EM, Wu, JH, Czerwinski, M, Sweet, CW, Wu, A, Tsai, YH, et al.. Differentiation of human intestinal organoids with endogenous vascular endothelial cells. Dev Cell 2020;54:516–28 e7. https://doi.org/10.1016/j.devcel.2020.07.023.Search in Google Scholar PubMed PubMed Central
154. Gjorevski, N, Nikolaev, M, Brown, TE, Mitrofanova, O, Brandenberg, N, DelRio, FW, et al.. Tissue geometry drives deterministic organoid patterning. Science 2022;375:eaaw9021. https://doi.org/10.1126/science.aaw9021.Search in Google Scholar PubMed PubMed Central
155. Frum, T, Spence, JR. hPSC-derived organoids: models of human development and disease. J Mol Med 2021;99:463–73. https://doi.org/10.1007/s00109-020-01969-w.Search in Google Scholar PubMed PubMed Central
156. Dye, BR, Hill, DR, Ferguson, MA, Tsai, YH, Nagy, MS, Dyal, R, et al.. In vitro generation of human pluripotent stem cell derived lung organoids. Elife 2015;4:e05098. https://doi.org/10.7554/elife.05098.Search in Google Scholar
157. Chen, YW, Huang, SX, de Carvalho, A, Ho, SH, Islam, MN, Volpi, S, et al.. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 2017;19:542–9. https://doi.org/10.1038/ncb3510.Search in Google Scholar PubMed PubMed Central
158. Spitalieri, P, Centofanti, F, Murdocca, M, Scioli, MG, Latini, A, Di Cesare, S, et al.. Two different therapeutic approaches for SARS-CoV-2 in hiPSCs-derived lung organoids. Cells 2022;11:1235. https://doi.org/10.3390/cells11071235.Search in Google Scholar PubMed PubMed Central
159. Kuhl, L, Graichen, P, von Daacke, N, Mende, A, Wygrecka, M, Potaczek, DP, et al.. Human lung organoids-a novel experimental and precision medicine approach. Cells 2023;12:2067. https://doi.org/10.3390/cells12162067.Search in Google Scholar PubMed PubMed Central
160. Lee, J, Kim, JH, Hong, SH, Yang, SR. Organoid model in idiopathic pulmonary fibrosis. Int J Stem Cells 2021;14:1–8. https://doi.org/10.15283/ijsc20093.Search in Google Scholar PubMed PubMed Central
161. Wilkinson, DC, Alva-Ornelas, JA, Sucre, JM, Vijayaraj, P, Durra, A, Richardson, W, et al.. Development of a three-dimensional bioengineering technology to generate lung tissue for personalized disease modeling. Stem Cells Transl Med 2017;6:622–33. https://doi.org/10.5966/sctm.2016-0192.Search in Google Scholar PubMed PubMed Central
162. Strikoudis, A, Cieslak, A, Loffredo, L, Chen, YW, Patel, N, Saqi, A, et al.. Modeling of fibrotic lung disease using 3D organoids derived from human pluripotent stem cells. Cell Rep 2019;27:3709–23 e5. https://doi.org/10.1016/j.celrep.2019.05.077.Search in Google Scholar PubMed PubMed Central
163. Assawachananont, J, Mandai, M, Okamoto, S, Yamada, C, Eiraku, M, Yonemura, S, et al.. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Rep 2014;2:662–74. https://doi.org/10.1016/j.stemcr.2014.03.011.Search in Google Scholar PubMed PubMed Central
164. Mandai, M, Fujii, M, Hashiguchi, T, Sunagawa, GA, Ito, SI, Sun, J, et al.. iPSC-Derived retina transplants improve vision in rd1 end-stage retinal-degeneration mice. Stem Cell Rep 2017;8:1112–3. https://doi.org/10.1016/j.stemcr.2017.03.024.Search in Google Scholar PubMed PubMed Central
165. Zhong, X, Gutierrez, C, Xue, T, Hampton, C, Vergara, MN, Cao, LH, et al.. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun 2014;5:4047. https://doi.org/10.1038/ncomms5047.Search in Google Scholar PubMed PubMed Central
166. Norrie, JL, Nityanandam, A, Lai, K, Chen, X, Wilson, M, Stewart, E, et al.. Retinoblastoma from human stem cell-derived retinal organoids. Nat Commun 2021;12:4535. https://doi.org/10.1038/s41467-021-24781-7.Search in Google Scholar PubMed PubMed Central
167. Roccio, M, Edge, ASB. Inner ear organoids: new tools to understand neurosensory cell development, degeneration and regeneration. Development 2019;146:dev177188. https://doi.org/10.1242/dev.177188.Search in Google Scholar PubMed PubMed Central
168. Liu, XP, Koehler, KR, Mikosz, AM, Hashino, E, Holt, JR. Functional development of mechanosensitive hair cells in stem cell-derived organoids parallels native vestibular hair cells. Nat Commun 2016;7:11508. https://doi.org/10.1038/ncomms11508.Search in Google Scholar PubMed PubMed Central
169. Koehler, KR, Mikosz, AM, Molosh, AI, Patel, D, Hashino, E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 2013;500:217–21. https://doi.org/10.1038/nature12298.Search in Google Scholar PubMed PubMed Central
170. Koehler, KR, Nie, J, Longworth-Mills, E, Liu, XP, Lee, J, Holt, JR, et al.. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat Biotechnol 2017;35:583–9. https://doi.org/10.1038/nbt.3840.Search in Google Scholar PubMed PubMed Central
171. Doda, D, Alonso Jimenez, S, Rehrauer, H, Carreno, JF, Valsamides, V, Di Santo, S, et al.. Human pluripotent stem cell-derived inner ear organoids recapitulate otic development in vitro. Development 2023;150:dev201865. https://doi.org/10.1242/dev.201865.Search in Google Scholar PubMed PubMed Central
172. Scherer, WF, Syverton, JT, Gey, GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med 1953;97:695–710. https://doi.org/10.1084/jem.97.5.695.Search in Google Scholar PubMed PubMed Central
173. Mirabelli, P, Coppola, L, Salvatore, M. Cancer cell lines are useful model systems for medical research. Cancers 2019;11:1098. https://doi.org/10.3390/cancers11081098.Search in Google Scholar PubMed PubMed Central
174. Yoshida, GJ. Applications of patient-derived tumor xenograft models and tumor organoids. J Hematol Oncol 2020;13:4. https://doi.org/10.1186/s13045-019-0829-z.Search in Google Scholar PubMed PubMed Central
175. Rygaard, J, Povsen, CO. Heterotransplantation of a human malignant tumour to “nude” mice. 1969. APMIS 2007;115:604–6. https://doi.org/10.1111/j.1600-0463.2007.apm_689a.x.Search in Google Scholar PubMed
176. Dietrich, C, Trub, A, Ahn, A, Taylor, M, Ambani, K, Chan, KT, et al.. INX-315, a selective CDK2 inhibitor, induces cell cycle arrest and senescence in solid tumors. Cancer Discov 2024;14:446–67. https://doi.org/10.1158/2159-8290.cd-23-0954.Search in Google Scholar PubMed PubMed Central
177. Hong, JH, Yong, CH, Heng, HL, Chan, JY, Lau, MC, Chen, J, et al.. Integrative multiomics enhancer activity profiling identifies therapeutic vulnerabilities in cholangiocarcinoma of different etiologies. Gut 2023;gutjnl-2023-330483. https://doi.org/10.1136/gutjnl-2023-330483.Search in Google Scholar PubMed
178. Li, Y, Guo, M, Qiu, Y, Li, M, Wu, Y, Shen, M, et al.. Autophagy activation is required for N6-methyladenosine modification to regulate ferroptosis in hepatocellular carcinoma. Redox Biol 2023;69:102971. https://doi.org/10.1016/j.redox.2023.102971.Search in Google Scholar PubMed PubMed Central
179. Xu, H, Jiao, D, Liu, A, Wu, K. Tumor organoids: applications in cancer modeling and potentials in precision medicine. J Hematol Oncol 2022;15:58. https://doi.org/10.1186/s13045-022-01278-4.Search in Google Scholar PubMed PubMed Central
180. Veninga, V, Voest, EE. Tumor organoids: opportunities and challenges to guide precision medicine. Cancer Cell 2021;39:1190–201. https://doi.org/10.1016/j.ccell.2021.07.020.Search in Google Scholar PubMed
181. Bian, S, Repic, M, Guo, Z, Kavirayani, A, Burkard, T, Bagley, JA, et al.. Genetically engineered cerebral organoids model brain tumor formation. Nat Methods 2018;15:631–9. https://doi.org/10.1038/s41592-018-0070-7.Search in Google Scholar PubMed PubMed Central
182. Hubert, CG, Rivera, M, Spangler, LC, Wu, Q, Mack, SC, Prager, BC, et al.. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res 2016;76:2465–77. https://doi.org/10.1158/0008-5472.can-15-2402.Search in Google Scholar PubMed PubMed Central
183. Wang, J, Li, X, Chen, H. Organoid models in lung regeneration and cancer. Cancer Lett 2020;475:129–35. https://doi.org/10.1016/j.canlet.2020.01.030.Search in Google Scholar PubMed
184. Zhang, H, Fillmore Brainson, C, Koyama, S, Redig, AJ, Chen, T, Li, S, et al.. Lkb1 inactivation drives lung cancer lineage switching governed by polycomb repressive complex 2. Nat Commun 2017;8:14922. https://doi.org/10.1038/ncomms14922.Search in Google Scholar PubMed PubMed Central
185. Dost, AFM, Moye, AL, Vedaie, M, Tran, LM, Fung, E, Heinze, D, et al.. Organoids model transcriptional hallmarks of oncogenic KRAS activation in lung epithelial progenitor cells. Cell Stem Cell 2020;27:663–78 e8. https://doi.org/10.1016/j.stem.2020.07.022.Search in Google Scholar PubMed PubMed Central
186. Kim, M, Mun, H, Sung, CO, Cho, EJ, Jeon, HJ, Chun, SM, et al.. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun 2019;10:3991. https://doi.org/10.1038/s41467-019-11867-6.Search in Google Scholar PubMed PubMed Central
187. Li, Z, Qian, Y, Li, W, Liu, L, Yu, L, Liu, X, et al.. Human lung adenocarcinoma-derived organoid models for drug screening. iScience 2020;23:101411. https://doi.org/10.1016/j.isci.2020.101411.Search in Google Scholar PubMed PubMed Central
188. Chen, JH, Chu, XP, Zhang, JT, Nie, Q, Tang, WF, Su, J, et al.. Genomic characteristics and drug screening among organoids derived from non-small cell lung cancer patients. Thorac Cancer 2020;11:2279–90. https://doi.org/10.1111/1759-7714.13542.Search in Google Scholar PubMed PubMed Central
189. Wang, HM, Zhang, CY, Peng, KC, Chen, ZX, Su, JW, Li, YF, et al.. Using patient-derived organoids to predict locally advanced or metastatic lung cancer tumor response: a real-world study. Cell Rep Med 2023;4:100911. https://doi.org/10.1016/j.xcrm.2022.100911.Search in Google Scholar PubMed PubMed Central
190. Zeng, X, Ma, Q, Li, XK, You, LT, Li, J, Fu, X, et al.. Patient-derived organoids of lung cancer based on organoids-on-a-chip: enhancing clinical and translational applications. Front Bioeng Biotechnol 2023;11:1205157. https://doi.org/10.3389/fbioe.2023.1205157.Search in Google Scholar PubMed PubMed Central
191. Chen, R, Lin, M, Gao, D. Major genomic mutations driving hepatocellular carcinoma. Genome Instab Dis 2023;4:15. https://doi.org/10.1007/s42764-023-00103-7.Search in Google Scholar
192. De Crignis, E, Hossain, T, Romal, S, Carofiglio, F, Moulos, P, Khalid, MM, et al.. Application of human liver organoids as a patient-derived primary model for HBV infection and related hepatocellular carcinoma. Elife 2021;10:e60747. https://doi.org/10.7554/elife.60747.Search in Google Scholar PubMed PubMed Central
193. Naruse, M, Masui, R, Ochiai, M, Maru, Y, Hippo, Y, Imai, T. An organoid-based carcinogenesis model induced by in vitro chemical treatment. Carcinogenesis 2020;41:1444–53. https://doi.org/10.1093/carcin/bgaa011.Search in Google Scholar PubMed
194. Sun, L, Wang, Y, Cen, J, Ma, X, Cui, L, Qiu, Z, et al.. Modelling liver cancer initiation with organoids derived from directly reprogrammed human hepatocytes. Nat Cell Biol 2019;21:1015–26. https://doi.org/10.1038/s41556-019-0359-5.Search in Google Scholar PubMed
195. Li, L, Knutsdottir, H, Hui, K, Weiss, MJ, He, J, Philosophe, B, et al.. Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight 2019;4:e121490. https://doi.org/10.1172/jci.insight.121490.Search in Google Scholar PubMed PubMed Central
196. Siegel, RL, Miller, KD, Jemal, A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7–30. https://doi.org/10.3322/caac.21387.Search in Google Scholar PubMed
197. Huang, L, Desai, R, Conrad, DN, Leite, NC, Akshinthala, D, Lim, CM, et al.. Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell Stem Cell 2021;28:1090–104 e6. https://doi.org/10.1016/j.stem.2021.03.022.Search in Google Scholar PubMed PubMed Central
198. Sachs, N, de Ligt, J, Kopper, O, Gogola, E, Bounova, G, Weeber, F, et al.. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 2018;172:373–86 e10. https://doi.org/10.1016/j.cell.2017.11.010.Search in Google Scholar PubMed
199. Mazzucchelli, S, Piccotti, F, Allevi, R, Truffi, M, Sorrentino, L, Russo, L, et al.. Establishment and morphological characterization of patient-derived organoids from breast cancer. Biol Proced Online 2019;21:12. https://doi.org/10.1186/s12575-019-0099-8.Search in Google Scholar PubMed PubMed Central
200. Li, X, Pan, B, Song, X, Li, N, Zhao, D, Li, M, et al.. Breast cancer organoids from a patient with giant papillary carcinoma as a high-fidelity model. Cancer Cell Int 2020;20:86. https://doi.org/10.1186/s12935-020-01171-5.Search in Google Scholar PubMed PubMed Central
201. Nayak, B, Balachander, GM, Manjunath, S, Rangarajan, A, Chatterjee, K. Tissue mimetic 3D scaffold for breast tumor-derived organoid culture toward personalized chemotherapy. Colloids Surf B Biointerfaces 2019;180:334–43. https://doi.org/10.1016/j.colsurfb.2019.04.056.Search in Google Scholar PubMed
202. Lindberg, K, Brown, ME, Chaves, HV, Kenyon, KR, Rheinwald, JG. In vitro propagation of human ocular surface epithelial cells for transplantation. Invest Ophthalmol Vis Sci 1993;34:2672–9.Search in Google Scholar
203. Rama, P, Matuska, S, Paganoni, G, Spinelli, A, De Luca, M, Pellegrini, G. Limbal stem-cell therapy and long-term corneal regeneration. N Engl J Med 2010;363:147–55. https://doi.org/10.1056/nejmoa0905955.Search in Google Scholar PubMed
204. Pellegrini, G, Traverso, CE, Franzi, AT, Zingirian, M, Cancedda, R, De Luca, M. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet 1997;349:990–3. https://doi.org/10.1016/s0140-6736(96)11188-0.Search in Google Scholar
205. Lancaster, MA, Knoblich, JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014;345:1247125. https://doi.org/10.1126/science.1247125.Search in Google Scholar PubMed
206. Eiraku, M, Sasai, Y. Self-formation of layered neural structures in three-dimensional culture of ES cells. Curr Opin Neurobiol 2012;22:768–77. https://doi.org/10.1016/j.conb.2012.02.005.Search in Google Scholar PubMed
207. Huch, M, Knoblich, JA, Lutolf, MP, Martinez-Arias, A. The hope and the hype of organoid research. Development 2017;144:938–41. https://doi.org/10.1242/dev.150201.Search in Google Scholar PubMed
208. Al Shihabi, A, Davarifar, A, Nguyen, HTL, Tavanaie, N, Nelson, SD, Yanagawa, J, et al.. Personalized chordoma organoids for drug discovery studies. Sci Adv 2022;8:eabl3674. https://doi.org/10.1126/sciadv.abl3674.Search in Google Scholar PubMed PubMed Central
209. Lensink, MA, Boers, SN, VA, MG, Jongsma, KR, Bredenoord, AL. Mini-gut feelings: perspectives of people with cystic fibrosis on the ethics and governance of organoid biobanking. Per Med 2021;18:241–54. https://doi.org/10.2217/pme-2020-0161.Search in Google Scholar PubMed
210. Boers, SN, de Winter-de Groot, KM, Noordhoek, J, Gulmans, V, van der Ent, CK, van Delden JJM, et al.. Mini-guts in a dish: perspectives of adult cystic fibrosis (CF) patients and parents of young CF patients on organoid technology. J Cyst Fibros 2018;17:407–15. https://doi.org/10.1016/j.jcf.2018.02.004.Search in Google Scholar PubMed
211. de Jongh, D, Massey, EK, consortium, V, Bunnik, EM, Lebreton, F, Bellofatto, K, et al.. Organoids: a systematic review of ethical issues. Stem Cell Res Ther 2022;13:337. https://doi.org/10.1186/s13287-022-02950-9.Search in Google Scholar PubMed PubMed Central
212. Munsie, M, Hyun, I, Sugarman, J. Ethical issues in human organoid and gastruloid research. Development 2017;144:942–5. https://doi.org/10.1242/dev.140111.Search in Google Scholar PubMed
213. Vyas, D, Baptista, PM, Brovold, M, Moran, E, Gaston, B, Booth, C, et al.. Self-assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology 2018;67:750–61. https://doi.org/10.1002/hep.29483.Search in Google Scholar PubMed PubMed Central
214. Takebe, T, Sekine, K, Kimura, M, Yoshizawa, E, Ayano, S, Koido, M, et al.. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep 2017;21:2661–70. https://doi.org/10.1016/j.celrep.2017.11.005.Search in Google Scholar PubMed
215. Karzbrun, E, Khankhel, AH, Megale, HC, Glasauer, SMK, Wyle, Y, Britton, G, et al.. Human neural tube morphogenesis in vitro by geometric constraints. Nature 2021;599:268–72. https://doi.org/10.1038/s41586-021-04026-9.Search in Google Scholar PubMed PubMed Central
216. Roth, JG, Brunel, LG, Huang, MS, Liu, Y, Cai, B, Sinha, S, et al.. Spatially controlled construction of assembloids using bioprinting. Nat Commun 2023;14:4346. https://doi.org/10.1038/s41467-023-40006-5.Search in Google Scholar PubMed PubMed Central
217. Qian, X, Nguyen, HN, Jacob, F, Song, H, Ming, GL. Using brain organoids to understand Zika virus-induced microcephaly. Development 2017;144:952–7. https://doi.org/10.1242/dev.140707.Search in Google Scholar PubMed PubMed Central
218. Li, H, Saucedo-Cuevas, L, Shresta, S, Gleeson, JG. The neurobiology of Zika virus. Neuron 2016;92:949–58. https://doi.org/10.1016/j.neuron.2016.11.031.Search in Google Scholar PubMed
219. Dekkers, JF, Berkers, G, Kruisselbrink, E, Vonk, A, de Jonge, HR, Janssens, HM, et al.. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med 2016;8:344ra84. https://doi.org/10.1126/scitranslmed.aad8278.Search in Google Scholar PubMed
220. Zhao, Z, Chen, X, Dowbaj, AM, Sljukic, A, Bratlie, K, Lin, L, et al.. Organoids. Nat Rev Methods Primers 2022;2:94. https://doi.org/10.1038/s43586-022-00174-y.Search in Google Scholar PubMed PubMed Central
221. Matsui, TK, Tsuru, Y, Hasegawa, K, Kuwako, KI. Vascularization of human brain organoids. Stem Cell 2021;39:1017–24. https://doi.org/10.1002/stem.3368.Search in Google Scholar PubMed
222. Mansour, AA, Goncalves, JT, Bloyd, CW, Li, H, Fernandes, S, Quang, D, et al.. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 2018;36:432–41. https://doi.org/10.1038/nbt.4127.Search in Google Scholar PubMed PubMed Central
223. Skylar-Scott, MA, Huang, JY, Lu, A, Ng, AHM, Duenki, T, Liu, S, et al.. Orthogonally induced differentiation of stem cells for the programmatic patterning of vascularized organoids and bioprinted tissues. Nat Biomed Eng 2022;6:449–62. https://doi.org/10.1038/s41551-022-00856-8.Search in Google Scholar PubMed PubMed Central
224. Homan, KA, Gupta, N, Kroll, KT, Kolesky, DB, Skylar-Scott, M, Miyoshi, T, et al.. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat Methods 2019;16:255–62. https://doi.org/10.1038/s41592-019-0325-y.Search in Google Scholar PubMed PubMed Central
225. Li, M, Izpisua Belmonte, JC. Organoids – preclinical models of human disease. N Engl J Med 2019;380:569–79. https://doi.org/10.1056/nejmra1806175.Search in Google Scholar PubMed
226. McCauley, HA, Wells, JM. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development 2017;144:958–62. https://doi.org/10.1242/dev.140731.Search in Google Scholar PubMed PubMed Central
227. Sasai, Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell 2013;12:520–30. https://doi.org/10.1016/j.stem.2013.04.009.Search in Google Scholar PubMed
228. Saw, TB, Doostmohammadi, A, Nier, V, Kocgozlu, L, Thampi, S, Toyama, Y, et al.. Topological defects in epithelia govern cell death and extrusion. Nature 2017;544:212–6. https://doi.org/10.1038/nature21718.Search in Google Scholar PubMed PubMed Central
229. Gupta, K, Ng, IC, Balachander, GM, Nguyen, BP, Tucker-Kellogg, L, Low, BC, et al.. Bile canaliculi contract autonomously by releasing calcium into hepatocytes via mechanosensitive calcium channel. Biomaterials 2020;259:120283. https://doi.org/10.1016/j.biomaterials.2020.120283.Search in Google Scholar PubMed
230. Warmflash, A, Sorre, B, Etoc, F, Siggia, ED, Brivanlou, AH. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods 2014;11:847–54. https://doi.org/10.1038/nmeth.3016.Search in Google Scholar PubMed PubMed Central
231. Kolesky, DB, Truby, RL, Gladman, AS, Busbee, TA, Homan, KA, Lewis, JA. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv Mater 2014;26:3124–30. https://doi.org/10.1002/adma.201305506.Search in Google Scholar PubMed
232. Huh, D, Matthews, BD, Mammoto, A, Montoya-Zavala, M, Hsin, HY, Ingber, DE. Reconstituting organ-level lung functions on a chip. Science 2010;328:1662–8. https://doi.org/10.1126/science.1188302.Search in Google Scholar PubMed PubMed Central
233. Condeelis, J, Pollard, JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006;124:263–6. https://doi.org/10.1016/j.cell.2006.01.007.Search in Google Scholar PubMed
234. Gjorevski, N, Sachs, N, Manfrin, A, Giger, S, Bragina, ME, Ordonez-Moran, P, et al.. Designer matrices for intestinal stem cell and organoid culture. Nature 2016;539:560–4. https://doi.org/10.1038/nature20168.Search in Google Scholar PubMed
235. Hendriks, D, Clevers, H, Artegiani, B. CRISPR-Cas tools and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell 2020;27:705–31. https://doi.org/10.1016/j.stem.2020.10.014.Search in Google Scholar PubMed
236. Nie, YZ, Zheng, YW, Miyakawa, K, Murata, S, Zhang, RR, Sekine, K, et al.. Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine 2018;35:114–23. https://doi.org/10.1016/j.ebiom.2018.08.014.Search in Google Scholar PubMed PubMed Central
237. Wang, S, Wang, X, Tan, Z, Su, Y, Liu, J, Chang, M, et al.. Human ESC-derived expandable hepatic organoids enable therapeutic liver repopulation and pathophysiological modeling of alcoholic liver injury. Cell Res 2019;29:1009–26. https://doi.org/10.1038/s41422-019-0242-8.Search in Google Scholar PubMed PubMed Central
238. Saborowski, A, Wolff, K, Spielberg, S, Beer, B, Hartleben, B, Erlangga, Z, et al.. Murine liver organoids as a genetically flexible system to study liver cancer in vivo and in vitro. Hepatol Commun 2019;3:423–36. https://doi.org/10.1002/hep4.1312.Search in Google Scholar PubMed PubMed Central
239. Li, Z, Araoka, T, Wu, J, Liao, HK, Li, M, Lazo, M, et al.. 3D culture supports long-term expansion of mouse and human nephrogenic progenitors. Cell Stem Cell 2016;19:516–29. https://doi.org/10.1016/j.stem.2016.07.016.Search in Google Scholar PubMed PubMed Central
240. Taguchi, A, Kaku, Y, Ohmori, T, Sharmin, S, Ogawa, M, Sasaki, H, et al.. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 2014;14:53–67. https://doi.org/10.1016/j.stem.2013.11.010.Search in Google Scholar PubMed
241. Takasato, M, Er, PX, Becroft, M, Vanslambrouck, JM, Stanley, EG, Elefanty, AG, et al.. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol 2014;16:118–26. https://doi.org/10.1038/ncb2894.Search in Google Scholar PubMed
242. Huang, L, Holtzinger, A, Jagan, I, BeGora, M, Lohse, I, Ngai, N, et al.. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med 2015;21:1364–71. https://doi.org/10.1038/nm.3973.Search in Google Scholar PubMed PubMed Central
243. Huch, M, Bonfanti, P, Boj, SF, Sato, T, Loomans, CJ, van de Wetering, M, et al.. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 2013;32:2708–21. https://doi.org/10.1038/emboj.2013.204.Search in Google Scholar PubMed PubMed Central
244. Nakano, T, Ando, S, Takata, N, Kawada, M, Muguruma, K, Sekiguchi, K, et al.. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012;10:771–85. https://doi.org/10.1016/j.stem.2012.05.009.Search in Google Scholar PubMed
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Articles in the same Issue
- Frontmatter
- Editorial
- Opportunity and challenge of rehabilitation medicine in China
- Reviews
- Engineering pluripotent stem cells with synthetic biology for regenerative medicine
- Assembling the RNA therapeutics toolbox
- Organoids as preclinical models of human disease: progress and applications
- Perspectives
- The role of bile acids in human aging
- Hormone-based pharmacotherapy for metabolic dysfunction-associated fatty liver disease
- Letter
- Clinical characteristics and pharmacokinetics of PAXLOVID in COVID-19 patients with hematological tumor
Articles in the same Issue
- Frontmatter
- Editorial
- Opportunity and challenge of rehabilitation medicine in China
- Reviews
- Engineering pluripotent stem cells with synthetic biology for regenerative medicine
- Assembling the RNA therapeutics toolbox
- Organoids as preclinical models of human disease: progress and applications
- Perspectives
- The role of bile acids in human aging
- Hormone-based pharmacotherapy for metabolic dysfunction-associated fatty liver disease
- Letter
- Clinical characteristics and pharmacokinetics of PAXLOVID in COVID-19 patients with hematological tumor

