Abstract
Ecological studies and epidemiology need to use group averaged data to make inferences about individual patterns. However, using correlations based on averages to estimate correlations of individual scores is subject to an “ecological fallacy”. The purpose of this article is to create distributions of Pearson R correlation values computed from grouped averaged or aggregate data using Monte Carlo simulations and random sampling. We show that, as the group size increases, the distributions can be approximated by a generalized hypergeometric distribution. The expectation of the constructed distribution slightly underestimates the individual Pearson R value, but the difference becomes smaller as the number of groups increases. The approximate normal distribution resulting from Fisher’s transformation can be used to build confidence intervals to approximate the Pearson R value based on individual scores from the Pearson R value based on the aggregated scores.
1 Introduction
The relationship between the Pearson R and regression coefficients computed from individual scores and the Pearson R and regression coefficients computed from grouped averaged or aggregate scores has been the subject of many papers [4, 6, 8]. Ecological studies and epidemiology often need to use group averaged data to make inferences about individual patterns [14]. However, using correlations based on averages to estimate correlations of individual scores is subject to an “ecological fallacy”. Robinson [13] demonstrated that the correlation of averages generally does not agree with the correlation of the original scores.
Knapp [9] relates within-aggregate (i.e. within group) correlations
where
Our objective is to describe the distribution of the Pearson
2 Distribution of Pearson R coefficient computed from a sample
2.1 Sampling from a bivariate distribution
If one samples
where ρ is the level of correlation,
will have the distribution
where Γ is the gamma function and
Moreover, the estimate provided by the sample is biased and underestimates ρ.
The expectation
where
The distribution (2.2) is not symmetric. However, Fisher [2] proposed the transformation
and it can be shown [10] that the resulting distribution approaches a normal distribution with standard deviation
as
2.2 Bivariate distribution of averages
The distribution of averages sampled from a bivariate distribution (2.1)
is described by [3] as
where

The analytical distribution (2.2) accurately represents the distribution created by n groups, where each of the n groups is formed by averaging m scores sampled from a bivariate distribution with
Figure 1 confirms this with a Monte Carlo simulation and random sampling for
3 Distribution of Pearson R x ¯ , y ¯
3.1 Defining a partition
We now describe the distribution of Pearson R coefficients created from group averages formed from a fixed sample of scores of length n. This requires us to define a partition of a set of original scores, where the scores are divided into subsets of equal size m.
Given a set of original scores
where each index
and so the union of all the
There exist
groups
Assemble all the pairs
Let us illustrate the process with an actual example.
Suppose we have the following
The average
The number of partitions that can be constructed using groups of size m from n individual scores is
The expression in (3.3) is derived by first choosing m elements from n elements,
number of ways of choosing all groups.
However, since order does not matter when choosing a partition, equation (3.4) needs to be divided by the number of ways of reordering the groups (i.e.
Figure 2 shows the number of partitions for different group sizes m for

3.2 Constructing the Pearson R x ¯ , y ¯
Section 2.2 showed that the Pearson R coefficient generated from averaged data sampled from a bivariate distribution follows the distribution described by equation (2.2).
In contrast, to create the Pearson R distribution from a partition, a set of n individual scores is chosen and remains fixed.
Each partition rearranges the n original scores into
Constructing the exact distribution using partitions would require one to compute the fraction of all possible partitions that produce a Pearson
List of symbols.
| Pearson R based on individual scores | |
| Pearson R based on group averages computed from partition | |
| Average of | |
| ρ | Level of correlation from bivariate distribution |
| n | Number of original scores |
| m | Size of groups |
| Number of groups |
We begin by generating the scores
where each
In Figure 3, we sample 1,000 different sets of

Difference between

Difference between

Difference in maximum and minimum value
To address the impact a specific individual sample has on the set of
Equation (3.7) is generated by replacing ρ with
Figure 6 compares the analytical distribution
to the partition distribution formed by randomly generating 50,000 partitions for

Comparing the analytical distribution (3.8) to the partition distribution for

Comparing the analytical distribution (3.8) to the partition distribution for

Difference between analytical distribution (3.8) and partition distribution for
We also create the partition distribution for scores randomly sampled from a uniform distribution,

Difference between analytical distribution (3.8) and partition distribution for
Figure 10 uses different group sizes

Comparing the analytical distribution (3.8) to the partition distribution for number of groups

Comparing the analytical distribution to the partition distribution for different values of ρ,

Comparing the analytical distribution to the partition distribution for different values of ρ for
Figure 12 compares the analytical distribution described by (3.8) to the partition distribution formed by randomly generating 50,000 partitions for
Figure 13 shows the ratio
of the standard deviation of the analytical distribution
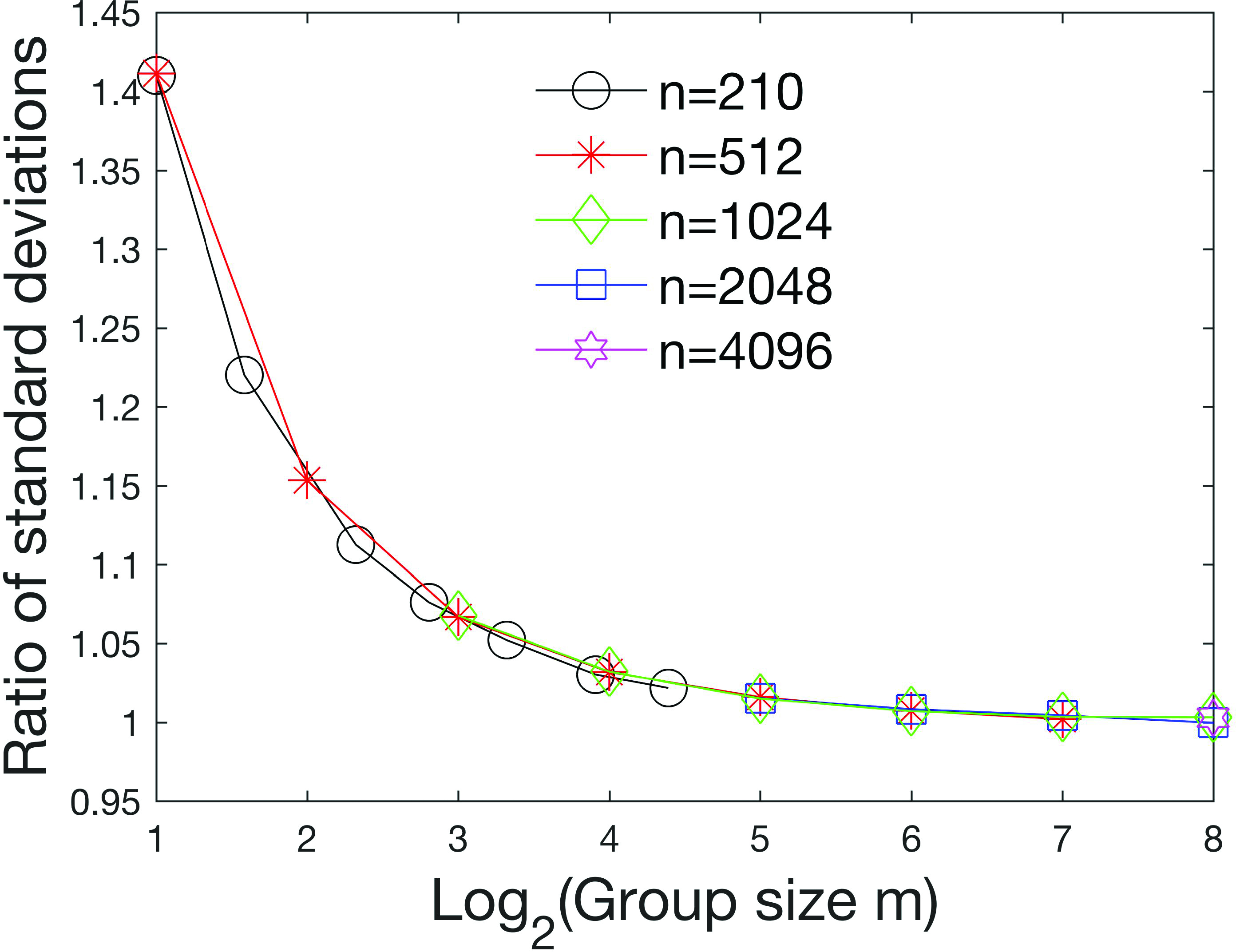
Ratio of standard deviation of analytical distribution to the partition distribution (3.9) for
The difference between the maximum ratio and the minimum ratio over all 100 samples for all values of n and m is plotted in Figure 14.
For some sample sizes (e.g.
These Monte Carlo simulations suggest that the distribution of Pearson
4 Constructing confidence intervals
4.1 Confidence intervals for large group sizes m
Since the partition distribution will approach (3.8) as the group size m increases, Fisher’s transformation (2.5) can be used to transform the set of
For a C % confidence interval,
For example, for a 95 % confidence interval, the value of
where
to generate the confidence interval
Equations (4.1)–(4.4) can be used to generate confidence intervals
95 % confidence intervals
| 20 | |||||
| 40 | |||||
| 80 | |||||
| 160 | |||||
| 320 |

Width of confidence interval
4.2 Confidence intervals for small group sizes m ≤ 8
If the group sizes m are not large, the process of generating the confidence intervals shown in equations (4.1)–(4.4) will need to be revised to account for the fact that the analytical distribution has a larger standard deviation compared to the partition distribution.
Figure 16 compares the analytical distribution (3.8) to the partition distribution created through Monte Carlo simulation and random sampling after Fisher’s transformation (2.5) is applied for

Comparing the analytical distribution (3.8) to the partitioning distribution for different group sizes m for
We assess whether the partition distributions for small group sizes
In (4.5), i refers to the location of the Fisher z-value when all the values are arranged in ascending order, and I refers to the total number of values in the distribution.
If the plot of the Fisher z-values versus the z-values extracted from the normal distribution is linear, the partition distribution is normal [15].
This plot is shown in Figure 17.
Except for extreme z-values,

The z-values extracted from a normal distribution are plotted against the Fisher z-values for group sizes
Figure 18 plots the ratio
of the standard deviation of Fisher’s transformed analytical distribution
to the standard deviation of Fisher’s transformed partition distribution
Figure 18 only uses combinations of n and m, so the number of groups
Equations (4.1)–(4.4) can still be used to compute the confidence intervals, but equation (4.3) is replaced with
(4.9)
We also increase the standard deviation by 3 % (reflected by the 1.03 in equation (4.9)) to account for the bias (3.7) and the range in standard deviations seen in Figure 14.
4.3 Assessing the accuracy of the confidence intervals
A simulation is performed to assess the accuracy of the confidence intervals computed using (4.9).
A value of ρ is selected from
Simulation to assess the validity of the confidence intervals (4.9) for different group sizes m and different values of ρ.
Each value of m and ρ shows the minimum and maximum percentage of
| m | |||||
| 2 | |||||
| 4 | |||||
| 8 | |||||
| 16 |
5 Conclusion
We have constructed the partition distribution of the Pearson
Funding source: National Institute of General Medical Sciences
Award Identifier / Grant number: P20GM103451
Funding statement: This is research is supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103451. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
Acknowledgements
Thank you to Ana Vasilic and Jose Pacheco for their valuable insights and feedback.
References
[1] G. Firebaugh, A rule for inferring individual-level relationships from aggregate data, Amer. Sociological Rev. 43 (1978), no. 4, 522–557. 10.2307/2094779Search in Google Scholar
[2] R. A. Fisher, On the probable error of a coefficient of correlation deduced from a small sample, Metron 1 (1921), 3–32. Search in Google Scholar
[3] H. Gatignon, Statistical Analysis of Management Data, 2nd ed., Springer, New York, 2010. 10.1007/978-1-4419-1270-1Search in Google Scholar
[4] L. A. Goodman, Ecological regressions and the behavior of individuals, Amer. Sociological Rev. 18 (1953), 663–664. 10.2307/2088121Search in Google Scholar
[5] M. T. Hannan and L. Burstein, Estimation from grouped observations, Amer. Sociological Rev. 39 (1974), no. 3, 374–392. 10.2307/2094296Search in Google Scholar
[6] J. W. Halliwell, Dangers inherent in correlating averages, J. Educational Res. 55 (1962), no. 7, 327–329. 10.1080/00220671.1962.10882826Search in Google Scholar
[7] J. L. Hammond, Two sources of error in ecological correlations, Amer. Sociological Rev. 38 (1973), no. 6, 764–777. 10.2307/2094137Search in Google Scholar
[8] L. Irwin and A. J. Lichtman, Across the great divide: Inferring individual level behavior from aggregate data, Polit. Methodology 3 (1976), 411–439. Search in Google Scholar
[9] T. R. Knapp, The unit-of-analysis problem in applications of simple correlation analysis to educational research, J. Educational Stat. 2 (1977), no. 3, 171–186. 10.3102/10769986002003171Search in Google Scholar
[10] R. J. Muirhead, Aspects of Multivariate Statistical Theory, John Wiley & Sons, Hoboken, 2005. Search in Google Scholar
[11] C. Ostroff, Comparing correlations based on individual-level and aggregated data, J. Appl. Psychol. 78 (1993), no. 4, 569–582. 10.1037/0021-9010.78.4.569Search in Google Scholar
[12] S. Piantasosi, D. P. Byar and S. B. Green, The ecological fallacy, Amer. J. Epidemiology 127 (1998), no. 6, 893–904. 10.1093/oxfordjournals.aje.a114892Search in Google Scholar PubMed
[13] W. S. Robinson, Ecological correlations and the behavior of individuals, Amer. Sociological Rev. 15 (1950), 351–357. 10.2307/2087176Search in Google Scholar
[14] S. Swhwartz, The fallacy of the ecological fallacy: The potential misuse of a concept and the consequences, Amer. J. Public Health 84 (1994), no. 5, 819–824. 10.2105/AJPH.84.5.819Search in Google Scholar
[15] M. Sullivan, Fundamentals of Statistics. Informed Decision Using Data, 5th ed., Pearson, Boston, 2018. Search in Google Scholar
[16] T. Vesala, U. Rannik, M. Leclerc, T. Foken and K. Sabelfeld, Flux and concentration footprints, Agricultural Forest Meteorol. 127 (2004), no. 3–4, 111–116. 10.1016/j.agrformet.2004.07.007Search in Google Scholar
© 2020 Walter de Gruyter GmbH, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 Public License.
Articles in the same Issue
- Frontmatter
- Why simple quadrature is just as good as Monte Carlo
- Describing the Pearson 𝑅 distribution of aggregate data
- Approximation of Euler–Maruyama for one-dimensional stochastic differential equations involving the maximum process
- A Bayesian inference for the penalized spline joint models of longitudinal and time-to-event data: A prior sensitivity analysis
- A Bayesian procedure for bandwidth selection in circular kernel density estimation
Articles in the same Issue
- Frontmatter
- Why simple quadrature is just as good as Monte Carlo
- Describing the Pearson 𝑅 distribution of aggregate data
- Approximation of Euler–Maruyama for one-dimensional stochastic differential equations involving the maximum process
- A Bayesian inference for the penalized spline joint models of longitudinal and time-to-event data: A prior sensitivity analysis
- A Bayesian procedure for bandwidth selection in circular kernel density estimation




