Gastroesophageal reflux disease influences blood pressure components, lipid profile and cardiovascular diseases: Evidence from a Mendelian randomization study
-
Qiang Wu
, Dazhi Lan
and Qiang Su
Abstract
Background
Gastroesophageal reflux disease (GERD) is a prevalent gastrointestinal disorder associated with a range of cardiovascular and metabolic complications. However, the relationship between GERD and blood pressure components, lipid profile, and cardiovascular diseases remains unclear.
Methods
Leveraging genetic variants associated with GERD as instrumental variables, we performed this Mendelian randomization (MR) analyses. Blood pressure components, lipid profile parameters, as well as cardiovascular diseases were considered as outcomes. Furthermore, we conducted reverse MR analysis to explore the association of these factors with the risk of GERD.
Results
Our MR analysis discovered a potential causal influence of GERD on blood pressure components, with genetically predicted GERD positively associated with systolic blood pressure (β = 0.053, P = 0.036), diastolic blood pressure (β = 0.100, P < 0.001), and mean arterial pressure (β = 0.106, P < 0.001). Additionally, genetically predicted GERD showed a significant impact on lipid profile, leading to increased genetically predicted levels of low-density lipoprotein (LDL) cholesterol (β = 0.093, P < 0.001), and triglycerides (β = 0.153, P < 0.001), while having a negative effect on high-density lipoprotein (HDL) cholesterol (β = -0.115, P = 0.002). Furthermore, our study indicated a noteworthy causal association between genetically predicted GERD and increased risk of myocardial infarction [odds ratio (OR) = 1.272, P = 0.019)] and hypertension (OR = 1.357, P < 0.001). No significant association was found between GERD and pulse pressure, total cholesterol, heart failure, and atrial fibrillation (P > 0.05). Reverse MR analysis indicates that blood pressure components, lipid profile, and cardiovascular diseases do not lead to an increased risk of GERD (all P > 0.05). Furthermore, mediation MR analysis reveals that LDL cholesterol (proportion mediated: 19.99%, 95% CI: 4.49% to 35.50%), HDL cholesterol (proportion mediated: 11.71%, 95% CI: 5.23% to 18.19%), and hypertension (proportion mediated: 35.09%, 95% CI: 24.66% to 45.53%) mediated the effect of GERD on myocardial infarction, while other factors did not participate in this pathway.
Conclusions
This MR study provides evidence supporting a causal relationship between GERD and alterations in blood pressure components, lipid profile, and increased risk of cardiovascular diseases.
Introduction
Gastroesophageal reflux disease (GERD),[1,2,3] a common gastrointestinal disorder, has been traditionally associated with symptoms such as heartburn and regurgitation. However, emerging evidence suggests that the impact of GERD extends beyond the confines of the esophagus, with potential implications for cardiovascular and metabolic health.[4,5,6] However, there remains controversy regarding the association between GERD and lipid profile and cardiovascular diseases. For instance, the study by Ha et al.[6] demonstrated that GERD patients have higher triglyceride levels compared to non-GERD patients, while no difference was found in total cholesterol and high-density lipoprotein (HDL) cholesterol. On the contrary, Kallel et al.[7] study found no significant difference in triglyceride between GERD patients and GERD-free patients. For cardiovascular disease, studies by Huang et al.[8] and Maret-Ouda et al.[9] elucidate the association between GERD and a high risk of atrial fibrillation. However, research by Bunch et al.[10] suggests that GERD does not lead to a high risk of atrial fibrillation.
Furthermore, it is essential to emphasize that conclusions from previous research are subject to potential confounding factors, such as age, body mass index (BMI), medication usage, etc. To address the aforementioned challenge, we conducted a Mendelian randomization (MR) study, leveraging genetic instruments associated with GERD as instrumental variables. In addition, it is well-known that MR provides a robust method to assess causality by utilizing genetic variants as proxies for exposures, mitigating issues related to confounding factors that commonly present in observational studies.[11,12] Thus, in this study, we perform an MR study aiming to elucidate the potential causal association between GERD and different outcomes, specifically focusing on blood pressure components [systolic blood pressure (SBP), diastolic blood pressure (DBP), pulse pressure (PP), and mean arterial pressure (MAP)], lipid profile parameters (low-density lipoprotein [LDL] cholesterol, HDL cholesterol, triglycerides, and total cholesterol), and disease risks including myocardial infarction, heart failure, atrial fibrillation, and hypertension.
Methods
Study design
We conducted a MR analysis utilizing large summary statistics from genome-wide association studies (GWAS) to investigate the association between GERD and blood pressure components, lipid profile, and disease risk. Furthermore, we conducted reverse MR analysis to explore the association of these factors with the risk of GERD. MR employs single nucleotide polymorphisms (SNPs) as instrumental variables to explore the causal impact of exposure on outcome. This method is robust against confounding factors since SNPs are randomly distributed during meiosis following Mendel’s laws. The rationality of the MR analysis relies on three important assumptions:[11,13] (1) a robust association between SNPs and the exposure; (2) the relationship between exposure and outcome remains unaffected by other confounding factors; and (3) SNPs associated with exposure exclusively influence outcomes through the exposure and are not influenced by alternative pathways. In this MR study, GERD serves as the exposure, while blood pressure components, lipid profile, and disease risk serve as outcomes. The blood pressure components encompass SBP, DBP, PP and MAP. The lipid profile includes LDL cholesterol, HDL cholesterol, triglycerides, and total cholesterol. Disease risk comprises myocardial infarction, heart failure, atrial fibrillation, and hypertension. The flowchart of this MR study is shown in Figure 1. The analytical process followed the guidelines outlined in the STROBE-MR guidelines.[14]

The flowchart of this Mendelian randomization study. BP components: blood pressure components; SBP: systolic blood pressure; DBP: diastolic blood pressure; PP: pulse pressure; MAP: mean arterial pressure; LDL cholesterol: low-density lipoprotein cholesterol; HDL cholesterol: high-density lipoprotein cholesterol; MR: Mendelian randomization; IVW: inverse variance weighted.
Ethical consideration
This MR study adhered to ethical standards in the utilization of genetic and phenotypic data. The summary-level data used were attained from publicly website, and ethical approval was not required as the data were de-identified and did not involve direct contact with study participants. The study design prioritized privacy and confidentiality, ensuring compliance with ethical guidelines governing the responsible use of genetic information. Additionally, transparency and adherence to established research ethics principles were maintained throughout the analysis and reporting process.
Data source
Data source for exposure
The large-scale GWAS data for GERD were derived from a study conducted by Ong et al.[15] In Ong et al.’s study,[15] a cohort of 602,604 participants of European descent was recruited, comprising 129,080 individuals diagnosed with GERD and 473,524 controls. Within this study, thorough identification and examination of 2,320,781 SNPs were conducted to investigate their potential associations with GERD.
Data source for different outcomes
The data for blood pressure components were sourced from two distinct studies. Information on SBP and DBP was extracted from the research conducted by Mbatchou et al.[16] which included over 380,000 individuals of European ancestry. For SBP, details were obtained from 385,798 individuals, encompassing 10,783,907 SNPs, while for DBP, data were sourced from 385,801 individuals, involving 10,783,908 SNPs. PP and MAP data were drawn from the study conducted by Sakaue et al.[17] which recruited over 360,000 individuals of European descent. Specifically, PP data were derived from 360,863 individuals, covering 19,047,322 SNPs, and MAP data were sourced from 360,863 individuals, involving 19,053,944 SNPs.
The data for the lipid profile came from four different studies. LDL cholesterol information was taken from the study led by Klimentidis et al.[18] which had 431,167 participants of European descent and involved 16,293,344 SNPs. HDL cholesterol details were sourced from the research conducted by Mbatchou et al.[16] which included 357,810 participants of European descent and had 10,783,660 SNPs. Triglycerides data were extracted from the study by Richardson et al.[19] which enrolled 441,016 participants of European descent and included 12,321,875 SNPs. Total cholesterol data were obtained from the study conducted by Sakaue et al.[17] involving 344,278 participants of European descent and encompassing 19,043,498 SNPs.
The data on cardiovascular diseases also come from four distinct studies. Myocardial infarction data were extracted from the study led by Sakaue et al.[17] involving 461,823 participants of European descent. In this study, 20,917 individuals were diagnosed with myocardial infarction, while 440,906 were non-myocardial infarction cases, covering a total of 24,172,914 SNPs. Heart failure data were gathered from the research conducted by Shah et al.[20] which included 977,323 participants of European ancestry. Among them, 47,309 individuals were diagnosed with heart failure, and 930,014 were non-heart failure cases, with a total of 7,773,021 SNPs. Atrial fibrillation data were acquired from the study by Nielsen et al.[21] comprising 1,030,836 participants of European descent. In this cohort, 60,620 individuals were diagnosed with atrial fibrillation, and 970,216 were non-atrial fibrillation cases, involving a total of 33,519,037 SNPs. Hypertension GWAS data originated from the FinnGen dataset (ID number: finn-b-I9_HYPTENSESS_EXNONE), with 218,792 participants of European descent. Within this dataset, 42,857 individuals were diagnosed with hypertension, and 175,935 were non-hypertension cases, incorporating a total of 16,380,466 SNPs.
Selection of SNPs
The procedure of SNPs selection was carried out utilizing the aforementioned GWAS databases, adhering to the three crucial assumptions of MR. To mitigate potential issues associated with linkage disequilibrium, a rigorous clustering procedure was performed, using clustering windows with an r2 = 0.001 and a kb = 10000. Furthermore, a threshold of P < 5*e-08 was employed to detect SNPs drastically associated with GERD. Additionally, an examination for potential confounding factors was conducted on the identified SNPs. Confounding factors encompassed age, gender, BMI, obesity, smoking, alcohol consumption, waist circumference, hip circumference, waist-hip ratio, depression, anxiety, HbA1c levels, blood glucose, tumors, diabetes, stress, obstructive sleep apnea, chronic kidney disease, multiple sclerosis, rheumatoid arthritis, potential diseases, and medication usage. SNPs associated with these confounding factors will be excluded from this MR study. The association between SNPs and confounding factors was assessed using the online platform PhenoScanner[22] (http://www.phenoscanner.medschl.cam.ac.uk/), GWAS Catalog (https://www.ebi.ac.uk/gwas/), IEU OpenGWAS project (https://gwas.mrcieu.ac.uk/). Additionally, to exclude weak instrumental variables, the calculation of the F-statistic was performed. Instrumental variables that yielded an F-statistic of less than 10 were deemed weak and thus excluded from this study. Following previous literature,[23,24] the formula for calculating the F-statistic is: F = (beta/se)2.
Statistical analysis
In this study, we initially employed five different methods, including MR Egger, Weighted median, Inverse variance weighted (IVW), Simple mode, and Weighted mode, to explore the potential causal relationship between GERD and various outcomes. In addition, we utilized reverse MR analysis to investigate the effects of blood pressure components, lipid profile, and cardiovascular diseases on GERD. Among these, the IVW method was considered the primary analysis,[25,26] while the other four methods were regarded as secondary analyses. In addition, we calculated Q statistics to assess the heterogeneity among the included instrumental variables, where a P-value of less than 0.05 indicated the presence of significant heterogeneity. In our study analysis, we employed random-effects IVW analysis when encountering significant heterogeneity (P < 0.05), and fixed-effects IVW analysis was used in cases where significant heterogeneity was absent (P > 0.05). For the sensitivity analysis, we applied three different methods, including Weighted median, MR-Egger, and MR-PRESSO,[27] to assess the consistency of associations and address potential horizontal pleiotropy. We employed the Weighted median method[28] to evaluate potential biases deriving from invalid instruments. If over 50% of the weight in the meta-analysis is based on valid SNPs, this method yields dependable estimates. Furthermore, MR-Egger[29] was utilized to detect any potential directional pleiotropy. The p-value calculated in the MR Egger analysis is utilized to evaluate the presence of horizontal pleiotropy. A P-value < 0.05 indicates significant horizontal pleiotropy, while a P-value > 0.05 suggests the absence of significant horizontal pleiotropy. Additionally, in this study, MR-PRESSO analysis was also conducted to detect and address potential outlier SNPs. SNPs identified as outliers through this process will be removed from the study.
Univariable and multivariable MR analysis
In this analysis, we considered cardiovascular diseases significantly associated with GERD (P < 0.05) as the outcomes, with blood pressure components and lipid profile significantly associated with GERD (P < 0.05) as exposures. Initially, we conducted univariable MR analysis to explore the effects of blood pressure components and lipid profile on cardiovascular diseases. Blood pressure components and lipid profile that exhibited significant associations with cardiovascular diseases were subsequently included in the multivariable MR analysis to further estimate the independent effects of each exposure on cardiovascular diseases.
Mediation MR analysis
In the mediation MR analysis, we employed a two-step MR approach to investigate whether blood pressure components and lipid profile mediate the effect of GERD on cardiovascular diseases. In this process, GERD was considered as the exposure, while the blood pressure components and lipid profile significantly associated with cardiovascular diseases in the multivariable MR analysis were considered as mediators, with cardiovascular diseases as the outcome. The effect of GERD on blood pressure components and lipid profile was denoted as β1, and the effect of blood pressure components and lipid profile on cardiovascular diseases was denoted as β2. The total effect of GERD on cardiovascular diseases was denoted as β (total). The proportion of mediation by each blood pressure component in the association between GERD and cardiovascular diseases was calculated as β1 * β2 / β (total). The 95% confidence interval (CI) for the mediation effect was computed using the delta method.[30]
Within this study, results were depicted using β coefficients and 95% CI for continuous variables, while odds ratios (OR) and their corresponding 95% CI were employed for categorical variables. The data analysis for this MR investigation was carried out using RStudio software (version 4.2.2) along with the Mendelian Randomization, MRPRESSO, and TwoSampleMR packages. A P-value below 0.05 indicates significant statistical differences.
Results
Several large GWAS datasets used in this MR study were obtained from previous studies, all involving European populations. The populations included in different studies all had sample sizes exceeding 200,000 individuals, with the smallest sample size being 2l8,792 and the largest being l,030,836. In summary, characteristics of the GWAS datasets used in this MR study are provided in Table 1.
Characteristics of the GWAS datasets used in this mendelian randomization study
| Type | PMID/ID | Journal | Population | Year | Sample size (case/control) | Number of SNPs |
|---|---|---|---|---|---|---|
| Gastroesophageal reflux disease | 34187846 | Gut | European | 2021 | 602,604 (129,080/ 473,524) | 2,320,781 |
| Systolic blood pressure | 34017140 | Nat Genet | European | 2021 | 385,798 (-/-) | 10,783,907 |
| Diastolic blood pressure | 34017140 | Nat Genet | European | 2021 | 385,801 (-/-) | 10,783,908 |
| Pulse pressure | 34594039 | Nat Genet | European | 2021 | 360,863 (-/-) | 19,047,322 |
| Mean arterial pressure | 34594039 | Nat Genet | European | 2021 | 360,863 (-/-) | 19,053,944 |
| LDL cholesterol | 32493714 | Diabetes | European | 2020 | 431,167 (-/-) | 16,293,344 |
| HDL cholesterol | 34017140 | Nat Genet | European | 2021 | 357,810 (-/-) | 10,783,660 |
| Triglycerides | 32203549 | PLoS Med | European | 2020 | 441,016 (-/-) | 12,321,875 |
| Total cholesterol | 34594039 | Nat Genet | European | 2021 | 344,278 (-/-) | 19,043,498 |
| Myocardial infarction | 34594039 | Nat Genet | European | 2021 | 461,823 (20,917/440,906) | 24,172,914 |
| Heart failure | 31919418 | Nat Commun | European | 2020 | 977,323 (47,309/930,014) | 7,773,021 |
| Atrial fibrillation | 30061737 | Nat Genet | European | 2018 | 1,030,836 (60,620/970,216) | 33,519,037 |
| Hypertension | finn-b-I9_HYPTENSESS_EXNONE | - | European | 2021 | 218,792 (42,857/175,935) | 16,380,466 |
GWAS: genome-wide association studies; SNPs: single nucleotide polymorphisms; LDL cholesterol: low-density lipoprotein cholesterol; HDL cholesterol: high-density lipoprotein cholesterol.
Causal relationship between GERD and blood pressure components
In Figure 2, we explore the causal links between GERD and blood pressure components. In the final analysis, we identified a total of 8 SNPs associated with SBP, 8 SNPs with DBP, l2 SNPs with PP, and ll SNPs with MAP. Subsequent analyses using the IVW method revealed a significant causal relationship between genetically predicted GERD and SBP, DBP, and MAP. Genetically predicted GERD led to a significant increase in SBP (β = 0.053, 95% CI: 0.003 to 0.102, P = 0.036), DBP (β = 0.100, 95% CI: 0.045 to 0.156, P < 0.001), and MAP (β = 0.106, 95% CI: 0.055 to 0.157, P < 0.001). However, no significant association was observed between genetically predicted GERD and PP (β = 0.019, 95% CI: -0.031 to 0.068, P = 0.461), regardless of the method used (MR Egger, Weighted Median, IVW, Simple Mode, Weighted Mode, P > 0.05).

Causal relationship between gastroesophageal reflux disease and blood pressure components. MR: Mendelian randomization; SNP: single nucleotide polymorphism; CI: confidence intervals.
Causal relationship between GERD and lipid profile
Figure 3 illustrates the causal relationships between GERD and the lipid profile. A total of l3 SNPs were identified to be associated with LDL cholesterol, 8 SNPs with HDL cholesterol, 12 SNPs with triglycerides, and 12 SNPs with total cholesterol. Analyses using the IVW method indicated a significant causal relationship between genetically predicted GERD and LDL cholesterol, HDL cholesterol, and triglycerides. Genetically predicted GERD resulted in a significant increase in LDL cholesterol (β = 0.093, 95% CI: 0.052 to 0.135, P < 0.001) and triglycerides (β = 0.153, 95% CI: 0.113 to 0.192, P < 0.001), while causing a significant decrease in HDL cholesterol (β = -0.115, 95% CI: -0.187 to -0.042, P = 0.002). However, no significant association was found between genetically predicted GERD and total cholesterol (β = 0.015, 95% CI: -0.025 to 0.054, P = 0.474), regardless of the method used (P > 0.05 for each method).
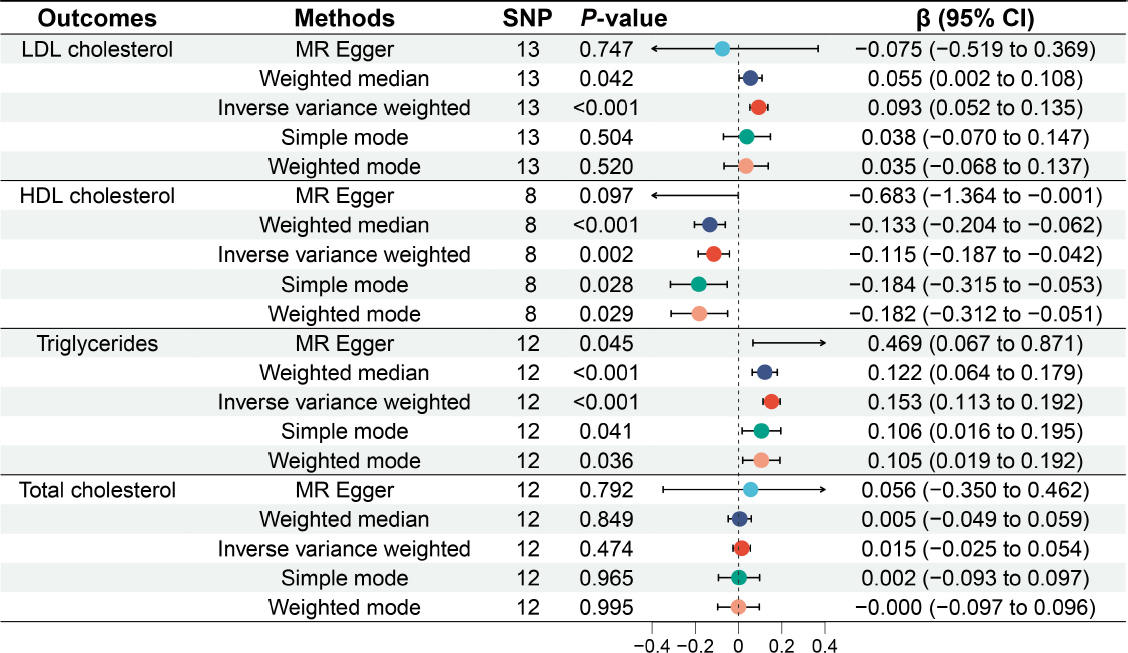
Causal relationship between gastroesophageal reflux disease and lipid profile. LDL cholesterol: low-density lipoprotein cholesterol; HDL cholestero high-density lipoprotein cholesterol; MR: Mendelian randomization; SNP: single nucleotide polymorphism; CI: confidence intervals.
Causal relationship between GERD and risk of cardiovascular diseases
Figure 4 outlines the relationships between GERD and disease risk. A total of 12 SNPs were associated with myocardial infarction, 8 SNPs with heart failure, 13 SNPs with atrial fibrillation, and 46 SNPs with hypertension. IVW analyses revealed a significant causal link between genetically predicted GERD and myocardial infarction, as well as hypertension. Genetically predicted GERD significantly increased the risk of myocardial infarction (OR = 1.272, 95% CI: 1.040 to 1.557, P = 0.019) and hypertension (OR = 1.357, 95% CI: 1.222 to 1.507, P < 0.001). Nevertheless, no significant association was detected between genetically predicted GERD and heart failure (OR = 1.174, 95% CI: 0.972 to 1.418, P = 0.095) or atrial fibrillation (OR = 1.134, 95% CI: 0.991 to 1.297, P = 0.067), suggesting that the presence of genetically predicted GERD does not drastically increase the risk of heart failure and atrial fibrillation.

Causal relationship between gastroesophageal reflux disease and cardiovascular diseases. OR: odds ratio; MR: Mendelian randomization; SNP: single nucleotide polymorphism; CI: confidence intervals.
Heterogeneity testing
We used two methods includin MR E er and IVW to evaluate the heterogeneity in the association between the exposure (GERD) and outcomes (SBP, DBP, PP, MAP, LDL cholesterol, HDL cholesterol, triglycerides, total cholesterol, myocardial infarction, heart failure, atrial fibrillation, hypertension). Detailed results are described in Table 2. Irrespective of the method used, whether MR Egger or IVW, we found no considerable heterogeneity in the relationship between GERD and SBP, DBP, MAP, LDL cholesterol, triglycerides, total cholesterol, myocardial infarction, heart failure, atrial fibrillation and hypertension (P > 0.05 for both methods). Additionally, MR Egger detected noteworthy heterogeneity in the relationship between GERD and PP (P = 0.036), while IVW suggested the relationship to be marginally significant (P = 0.050). Conversely, IVW detected considerable heterogeneity in the relationship between GERD and HDL cholesterol (P = 0.021), whereas MR Egger did not detect this significant heterogeneity (P = 0.077).
Heterogeneity test between gastroesophageal reflux disease and different outcomes
| Exposure | Outcomes | Methods | Q statistics | P-value |
|---|---|---|---|---|
| GRD | SBP | MR Egger | 5.851 | 0.440 |
| GRD | SBP | Inverse variance weighted | 6.373 | 0.497 |
| GRD | DBP | MR Egger | 7.747 | 0.257 |
| GRD | DBP | Inverse variance weighted | 7.870 | 0.344 |
| GRD | Pulse pressure | MR Egger | 19.379 | 0.036 |
| GRD | Pulse pressure | Inverse variance weighted | 19.659 | 0.050 |
| GRD | MAP | MR Egger | 15.044 | 0.090 |
| GRD | MAP | Inverse variance weighted | 15.440 | 0.117 |
| GRD | LDL cholesterol | MR Egger | 15.145 | 0.176 |
| GRD | LDL cholesterol | Inverse variance weighted | 15.914 | 0.195 |
| GRD | HDL cholesterol | MR Egger | 11.399 | 0.077 |
| GRD | HDL cholesterol | Inverse variance weighted | 16.515 | 0.021 |
| GRD | Triglycerides | MR Egger | 8.762 | 0.555 |
| GRD | Triglycerides | Inverse variance weighted | 11.164 | 0.430 |
| GRD | Total cholesterol | MR Egger | 9.415 | 0.493 |
| GRD | Total cholesterol | Inverse variance weighted | 9.456 | 0.580 |
| GRD | Myocardial infarction | MR Egger | 12.240 | 0.269 |
| GRD | Myocardial infarction | Inverse variance weighted | 13.610 | 0.255 |
| GRD | Heart failure | MR Egger | 3.918 | 0.688 |
| GRD | Heart failure | Inverse variance weighted | 4.793 | 0.685 |
| GRD | Atrial fibrillation | MR Egger | 12.268 | 0.344 |
| GRD | Atrial fibrillation | Inverse variance weighted | 13.854 | 0.310 |
| GRD | Hypertension | MR Egger | 58.765 | 0.067 |
| GRD | Hypertension | Inverse variance weighted | 59.871 | 0.068 |
GRD: gastroesophageal reflux disease; MR: Mendelian randomization; SBP: systolic blood pressure; DBP: diastolic blood pressure; MAP: mean arterial pressure; LDL cholesterol: low-density lipoprotein cholesterol; HDL cholesterol: high-density lipoprotein cholesterol; SNPs: single nucleotide polymorphisms; CI: confident interval.
Assessment of pleiotropy
To further investigate whether the relationship between GERD and SBP, DBP, PP, MAP, LDL cholesterol, HDL cholesterol, triglycerides, total cholesterol, myocardial infarction, heart failure, atrial fibrillation, and hypertension is influenced by other potential factors, we performed MR Egger regression to assess the presence of pleiotropy. The results displayed that each Egger intercept value was negligible (range from -0.034 to 0.026), with each P-value > 0.05, suggesting that the relationship between GERD and different outcomes is not affected by pleiotropy. Detailed results are provided in Table 3. Therefore, it can be concluded that the relationships between GERD and SBP, DBP, PP, MAP, LDL cholesterol, HDL cholesterol, triglycerides, total cholesterol, myocardial infarction, heart failure, atrial fibrillation, and hypertension are robust and not influenced by traditional confounding factors or potential confounders.
Assessment of pleiotropy between gastroesophageal reflux disease and different outcomes
| Exposure | Outcomes | Egger intercept | Se | P-value |
|---|---|---|---|---|
| Gastroesophageal reflux disease | Systolic blood pressure | -0.006 | 0.008 | 0.497 |
| Gastroesophageal reflux disease | Diastolic blood pressure | -0.003 | 0.010 | 0.768 |
| Gastroesophageal reflux disease | Pulse pressure | -0.003 | 0.008 | 0.712 |
| Gastroesophageal reflux disease | Mean arterial pressure | 0.004 | 0.008 | 0.638 |
| Gastroesophageal reflux disease | Low density lipoprotein cholesterol | 0.005 | 0.007 | 0.470 |
| Gastroesophageal reflux disease | High density lipoprotein cholesterol | 0.018 | 0.011 | 0.152 |
| Gastroesophageal reflux disease | Triglycerides | -0.010 | 0.006 | 0.152 |
| Gastroesophageal reflux disease | Total cholesterol | -0.001 | 0.006 | 0.844 |
| Gastroesophageal reflux disease | Myocardial infarction | -0.034 | 0.032 | 0.315 |
| Gastroesophageal reflux disease | Heart failure | -0.029 | 0.031 | 0.386 |
| Gastroesophageal reflux disease | Atrial fibrillation | 0.026 | 0.022 | 0.258 |
| Gastroesophageal reflux disease | Hypertension | -0.009 | 0.010 | 0.367 |
Reverse MR analysis
We further conducted a reverse MR analysis, using blood pressure components, lipid profile, and cardiovascular diseases as exposures, and GERD as the outcome. The results of the reverse MR analysis indicate that there is no significant (all P > 0.05) genetic association between blood pressure components, lipid profile, and cardiovascular diseases and the risk of GERD (Figure 5 and Figure 6).
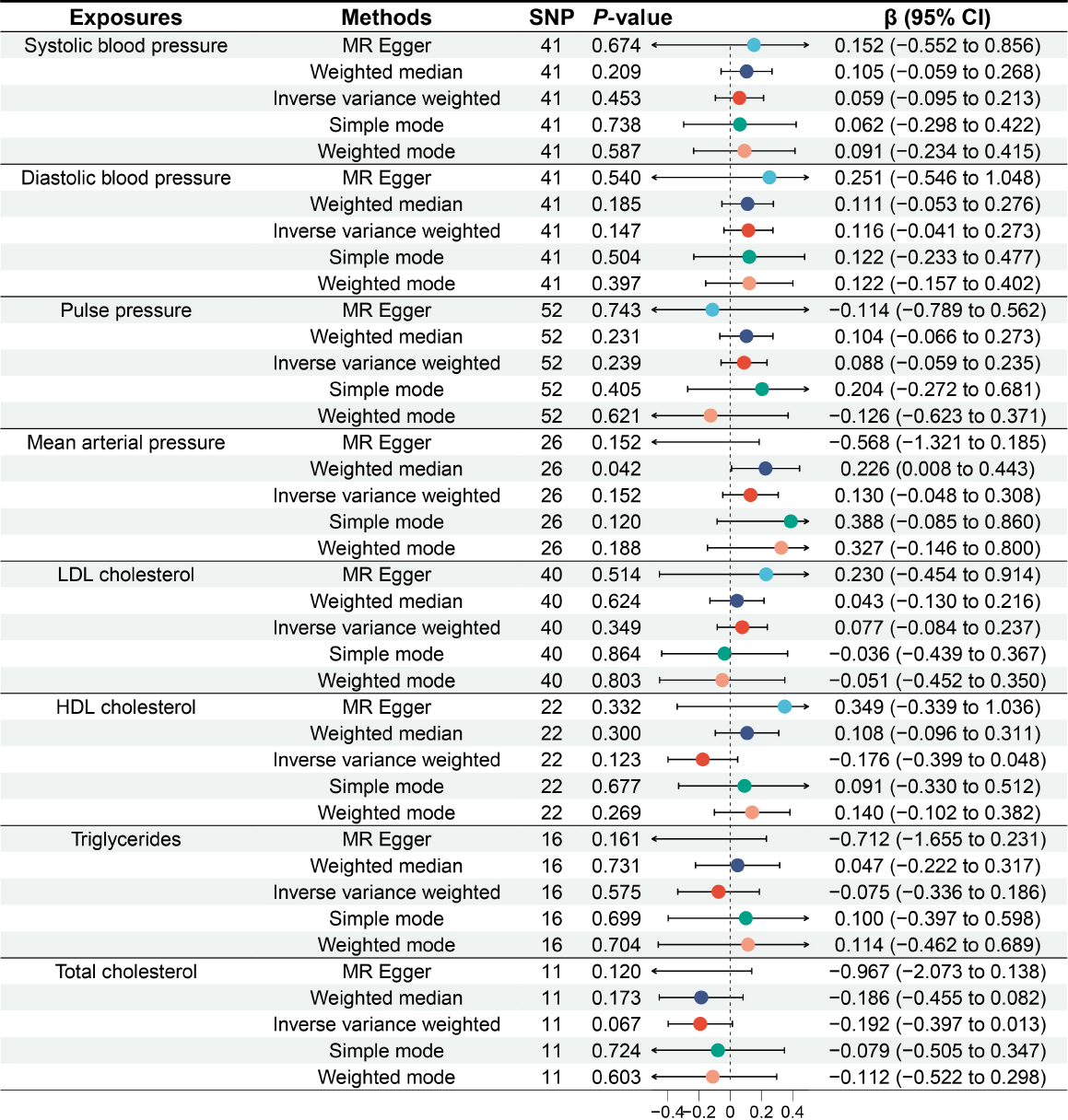
Causal relationship between blood pressure components, lipid profile and gastroesophageal reflux disease. MR: Mendelian randomization; SNP: single nucleotide polymorphism; CI: confidence intervals.

Causal relationship between cardiovascular diseases and gastroesophageal reflux disease. MR: Mendelian randomization; SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence intervals.
Univariable and multivariable MR analysis
We initially considered myocardial infarction as the outcome and other factors significantly associated with GERD as exposures (P < 0.05), using univariable and multivariable MR analysis to explore the relationship between myocardial infarction and these factors. Univariable MR analysis revealed that SBP (OR = 1.639, 95% CI: 1.355 to 1.983, P < 0.001), DBP (OR = 1.627, 95% CI: 1.399 to 1.893, P < 0.001), MAP (OR = 2.242, 95% CI: 1.805 to 2.785, P < 0.001), LDL cholesterol (OR = 1.678, 95% CI: 1.457 to 1.932, P < 0.001), HDL cholesterol (OR = 0.782, 95% CI: 0.694 to 0.882, P < 0.001), triglycerides (OR = 1.346, 95% CI: 1.094 to 1.658, P = 0.005), and hypertension (OR = 1.319, 95% CI: 1.152 to 1.509, P < 0.001) are associated with the risk of myocardial infarction. Detailed results are shown in Table 4. In the multivariable analysis, we found that LDL cholesterol (β = 1.748, 95% CI: 1.598 to 1.913, P < 0.001) and hypertension (OR = 1.355, 95% CI: 1.189 to 1.543, P < 0.001) are associated with an increased risk of myocardial infarction, while HDL cholesterol (OR = 0.807, 95% CI: 0.721 to 0.904, P < 0.001) is associated with a decreased risk of myocardial infarction (Table 5). We further explored the effects of LDL cholesterol, HDL cholesterol, triglycerides, and myocardial infarction on hypertension and found that LDL cholesterol (OR = 1.066, 95% CI: 0.924 to 1.229, P = 0.064), HDL cholesterol (OR = 0.900, 95% CI: 0.805 to 1.006, P = 0.062), triglycerides (OR = 1.170, 95% CI: 0.973 to 1.408, P = 0.096), and myocardial infarction (OR = 1.094, 95% CI: 0.994 to 1.204, P = 0.065) are not significantly associated with hypertension.
Results of univariable analysis associated with myocardial infarction
| Exposures | Methods | Number of SNP | OR | 95% CI | P-value |
|---|---|---|---|---|---|
| Systolic blood pressure | MR Egger | 133 | 1.655 | 0.714 to 3.837 | 0.243 |
| Systolic blood pressure | Weighted median | 133 | 1.633 | 1.332 to 2.002 | <0.001 |
| Systolic blood pressure | IVW | 133 | 1.639 | 1.355 to 1.983 | <0.001 |
| Systolic blood pressure | Simple mode | 133 | 1.259 | 0.739 to 2.146 | 0.398 |
| Systolic blood pressure | Weighted mode | 133 | 1.618 | 0.943 to 2.777 | 0.083 |
| Diastolic blood pressure | MR Egger | 118 | 1.284 | 0.683 to 2.416 | 0.439 |
| Diastolic blood pressure | Weighted median | 118 | 1.566 | 1.290 to 1.902 | <0.001 |
| Diastolic blood pressure | IVW | 118 | 1.627 | 1.399 to 1.893 | <0.001 |
| Diastolic blood pressure | Simple mode | 118 | 1.857 | 1.048 to 3.288 | 0.036 |
| Diastolic blood pressure | Weighted mode | 118 | 1.813 | 1.027 to 3.202 | 0.043 |
| Mean arterial pressure | MR Egger | 83 | 2.134 | 0.888 to 5.128 | 0.094 |
| Mean arterial pressure | Weighted median | 83 | 2.317 | 1.783 to 3.011 | <0.001 |
| Mean arterial pressure | IVW | 83 | 2.242 | 1.805 to 2.785 | <0.001 |
| Mean arterial pressure | Simple mode | 83 | 2.650 | 1.336 to 5.256 | 0.007 |
| Mean arterial pressure | Weighted mode | 83 | 2.611 | 1.354 to 5.036 | 0.005 |
| LDL cholesterol | MR Egger | 143 | 1.747 | 1.239 to 2.463 | 0.002 |
| LDL cholesterol | Weighted median | 143 | 1.674 | 1.381 to 2.029 | <0.001 |
| LDL cholesterol | IVW | 143 | 1.678 | 1.457 to 1.932 | <0.001 |
| LDL cholesterol | Simple mode | 143 | 2.034 | 1.264 to 3.273 | 0.004 |
| LDL cholesterol | Weighted mode | 143 | 1.742 | 1.295 to 2.344 | <0.001 |
| HDL cholesterol | MR Egger | 152 | 1.053 | 0.856 to 1.296 | 0.623 |
| HDL cholesterol | Weighted median | 152 | 0.878 | 0.749 to 1.028 | 0.106 |
| HDL cholesterol | IVW | 152 | 0.782 | 0.694 to 0.882 | <0.001 |
| HDL cholesterol | Simple mode | 152 | 0.789 | 0.467 to 1.334 | 0.378 |
| HDL cholesterol | Weighted mode | 152 | 0.911 | 0.780 to 1.064 | 0.241 |
| triglycerides | MR Egger | 64 | 2.163 | 1.272 to 3.678 | 0.006 |
| triglycerides | Weighted median | 64 | 1.455 | 1.112 to 1.904 | 0.006 |
| triglycerides | IVW | 64 | 1.346 | 1.094 to 1.658 | 0.005 |
| triglycerides | Simple mode | 64 | 1.509 | 0.863 to 2.639 | 0.154 |
| triglycerides | Weighted mode | 64 | 1.469 | 0.992 to 2.176 | 0.059 |
| Hypertension | MR Egger | 12 | 1.624 | 1.082 to 2.437 | 0.041 |
| Hypertension | Weighted median | 12 | 1.216 | 1.061 to 1.394 | 0.005 |
| Hypertension | IVW | 12 | 1.319 | 1.152 to 1.509 | <0.001 |
| Hypertension | Simple mode | 12 | 1.204 | 0.972 to 1.491 | 0.118 |
| Hypertension | Weighted mode | 12 | 1.212 | 1.041 to 1.412 | 0.031 |
MR: Mendelian randomization; LDL cholesterol: low-density lipoprotein cholesterol; HDL cholesterol: high-density lipoprotein cholesterol; SNP: single nucleotide polymorphisms; CI: confident interval.
Results of multivariable analysis related to myocardial infarction
| Exposures | Outcome | OR | 95% CI | P-value |
|---|---|---|---|---|
| LDL cholesterol | Myocardial infarction | 1.748 | 1.598 to 1.913 | <0.001 |
| HDL cholesterol | Myocardial infarction | 0.807 | 0.721 to 0.904 | <0.001 |
| Diastolic blood pressure | Myocardial infarction | 1.365 | 0.833 to 2.234 | 0.217 |
| Systolic blood pressure | Myocardial infarction | 0.781 | 0.474 to 1.285 | 0.330 |
| Mean arterial pressure | Myocardial infarction | 1.317 | 0.564 to 3.076 | 0.525 |
| Hypertension | Myocardial infarction | 1.355 | 1.189 to 1.543 | <0.001 |
| Triglycerides | Myocardial infarction | 0.886 | 0.785 to 1.001 | 0.053 |
LDL cholesterol: low-density lipoprotein cholesterol; HDL cholesterol: high-density lipoprotein cholesterol; OR: odds ratio; CI: confident interval.
Mediation MR analysis
We further employed mediation MR analysis to investigate how LDL cholesterol, HDL cholesterol, and hypertension mediate the effect of GERD on myocardial infarction. The results of the mediation MR analysis revealed that LDL cholesterol mediated 19.99% (95% CI: 4.49% to 35.50%) of the effect of GERD on myocardial infarction, HDL cholesterol mediated 11.71% (95% CI: 5.23% to 18.19%) of the effect of GERD on myocardial infarction, and hypertension mediated 35.09% (95% CI: 24.66% to 45.53%) of the effect of GERD on myocardial infarction. Detailed results are presented in Figure 7.

Mediation MR analysis. LDL cholesterol, HDL cholesterol and hypertension mediate the effects of gastroesophageal reflux disease on myocardial infarction. LDL cholesterol: low-density lipoprotein cholesterol; HDL cholesterol: high-density lipoprotein cholesterol.
Discussion
Key finding
This MR study provides novel evidence supporting a causal relationship between GERD and significant alterations in blood pressure components, lipid profile, and an increased risk of cardiovascular diseases. Specifically, genetically predicted GERD exhibited positive associations with SBP, DBP, and MAP. Furthermore, our MR analyses revealed a substantial impact of GERD on lipid profile parameters, with genetically predicted GERD being associated with elevated levels of LDL cholesterol and triglycerides, while concurrently showing a negative impact on HDL cholesterol. Furthermore, our study revealed a significant association between genetically predicted GERD and a high risk of myocardial infarction and hypertension. Additionally, mediation MR analysis elucidated that LDL cholesterol (19.99% [95% CI: 4.49% to 35.50%]), HDL cholesterol (11.71% [95% CI: 5.23% to 18.19%]), and hypertension (35.09% [95% CI: 24.66% to 45.53%]) mediate the effect of GERD on myocardial infarction. Nevertheless, no noteworthy associations were detected between genetically predicted GERD and PP, total cholesterol, heart failure, and atrial fibrillation.
GERD and blood pressure components
Presently, several prior studies have investigated the influence of GERD on blood pressure components. Ha et al.[6] conducted a study involving 150 participants diagnosed with GERD and 292 non-GERD individuals, revealing no significant differences in SBP (129.2 ± 1.4 vs. 130.7 ± 1.0 mmHg, P = 0.390) and DBP (78.4 ± 0.9 vs. 77.6 ± 0.6 mmHg, P = 0.437) between GERD patients and those without GERD. Similarly, Kallel et al.[7] included 54 participants diagnosed with GERD and 46 non-GERD participants, with no significant differences observed in SBP (122.07 ± 7.68 vs. 121.52 ± 12.9 mmHg, P = 0.16) and DBP (70.52 ± 12.04 vs. 70.23 ± 17.95 mmHg, P = 0.9) between GERD participants and those without GERD. Contrary to the studies performed by Ha[6] and Kallel,[7] Milovanovic et al.[31] found that GERD patients had higher SBP and PP compared to age- and gender-matched participants. Furthermore, based on a large-scale study, Kim et al.[32] found that GERD patients have higher SBP (121.0 [112.0–131.0 mmHg] ) compared to those without GERD (120.0 [110.0–130.0 mmHg]), while there was no significant difference in DBP between the two groups. It’s crucial to highlight that those previous studies had small sample sizes and couldn’t effectively account for confounding factors like age, gender, BMI, underlying diseases, or medication usage. In our study, utilizing MR analysis on a large-scale GWAS dataset, the presence of genetically predicted GERD significantly elevated SBP (β = 0.053, P = 0.036), DBP (β = 0.100, P < 0.001), and MAP (β = 0.106, P < 0.001), with no significant association observed with PP. The results of our study show different conclusions from previous research, likely attributed to the considerably larger sample size and the exclusion of potential confounding factors. For instance, participants with GERD are significantly older than non-GERD patients (44.59 ± 10.34 vs. 37.63 ± 14.41, P = 0.006) in Kallel’s study,[7] while those with GERD have a significantly lower BMI than non-GERD individuals (23.9 ± 0.3 vs. 24.8 ± 0.2, P = 0.026) in Ha’s study.[6] Thus, it is challenging to compare the differences in blood pressure components between GERD participants and non-GERD participants due to the potential influence of confounding factors. In our MR study, the influence of potential confounding factors has been eliminated, and the final conclusions are more accurate.
GERD and lipid profile
The association between GERD and lipid profile remains uncertain. In Ha et al. study,[6] a comparison of lipid profiles between GERD and non-GERD patients revealed a significant difference in triglycerides. GERD patients had significantly higher triglyceride levels than non-GERD counterparts (157.8 ± 19.5 vs. 120.5 ± 3.9 mg/dL, P = 0.013). However, there was no significant difference in total cholesterol and HDL cholesterol between GERD and non-GERD patients. Similarly, in Kallel et al. study,[7] a comparison of lipid profiles between GERD and nonGERD patients did not identify significant differences in total cholesterol (106.33 ± 61.6 vs. 90 ± 37.61 mg/dL, P = 0.14) and HDL cholesterol (47.38 ± 12.12 vs. 52.41 ± 22.02 mg/dL, P = 0.16). Additionally, in Kim et al. study,[32] the comparison of lipid profiles between GERD and non-GERD patients revealed lower LDL cholesterol and HDL cholesterol in GERD patients, along with higher total cholesterol and triglyceride levels compared to those without GERD. In contrast to prior studies, this MR study revealed noteworthy differences in lipid profiles between GERD and non-GERD patients. Specifically, genetically predicted GERD demonstrated a significant association with elevated LDL cholesterol (β = 0.093, P < 0.001) and triglycerides (β = 0.153, P < 0.001), alongside an association with reduced HDL cholesterol (β = -0.115, P = 0.002). However, no significant association was noticed between genetically predicted GERD and total cholesterol. The considerable sample size (> 200,000 individuals) and careful control for confounding factors in this MR study likely contribute to more stable results compared to previous research, as outlined above.
GERD and cardiovascular diseases
Nowadays, there are limited studies systematically assessing the impact of GERD on cardiovascular diseases. In a nationwide population-based study, Lei et al.[33] enrolled 54, 422 patients diagnosed with GERD and 269,572 age- and gender-matched controls. After a mean follow-up of 3.3 years, they identified a notable association between GERD and a high risk of myocardial infarction (hazard ratio [HR] = 1.48, 95% CI: 1.31 to 1.66, P < 0.001). In another large sample study, Eisa et al.[34] similarly noted an increased risk of myocardial infarction linked to the presence of GERD, and this association remained exist after adjusting for potential confounding factors. Aligning with prior research, our MR study also revealed a substantial association between genetically predicted GERD and an elevated risk of myocardial infarction (OR = 1.272, 95% CI: 1.040 to 1.557, P = 0.019). Presently, there is a lack of research directly evaluating the causative relationship between GERD and hypertension. In a study conducted by Li et al.,[35] 86 hypertensive patients were enrolled, revealing that 44.2% of those with hypertension also experienced GERD, indicating a notable association between hypertension and GERD. Another study [36] found that in hypertensive patients, the prevalence of silent GERD and GERD were 15.1% and 31.4%, respectively. However, while those studies suggest an association between GERD and hypertension, the potential causative relationship remains unclear. To our knowledge, the findings of our MR study are the first to reveal that genetically predicted GERD is associated with an increased risk of hypertension (OR = 1.357, 95% CI: 1.222 to 1.507, P < 0.001), systematically elucidating the causal relationship between GERD and hypertension.
This study performed by Sun et al.[37] is the initial investigation into the association between GERD and heart failure, and found that the presence of GERD is associated with a higher risk of heart failure. However, it needs to be emphasized that the study by Sun et al.[37] did not eliminate instrumental variables significantly associated with confounding factors, thereby leading to potential bias in the final conclusion. Contrary to the study by Sun et al.[37] our findings indicate that genetically predicted GERD is not significantly associated with a high risk of heart failure. One major reason attributing to the difference is that instrumental variables associated with confounding factors, such as age, gender, BMI, obesity, smoking, alcohol consumption, waist circumference, hip circumference, waist-hip ratio, depression, anxiety, HbA1c levels, blood glucose, tumors, diabetes, stress, obstructive sleep apnea, chronic kidney disease, multiple sclerosis, rheumatoid arthritis, potential diseases, and medication usage, were excluded from our study, whereas the exclusion of confounding factors is not mentioned in Sun’s study.[37] In that case, the conclusions of our study are more accurate because the influence of confounding factors on the findings can be considered negligible.
On the other hand, there is significant controversy regarding the causal relationship between GERD and atrial fibrillation. A study[8] with 29,688 diagnosed GERD patients and 29,597 non-GERD patients suggested that GERD independently contributes to the occurrence of atrial fibrillation (HR = 1.31, 95% CI: 1.06 to 1.61, P = 0.013). Another study provided evidence that the presence of GERD increases the risk of atrial fibrillation in individuals below the age of 60.[9] Similarly, Sun et al. study also found that the presence of GERD also significantly increased the risk of atrial fibrillation. In contrast, Bunch et al. study[10] found no association between the presence of any GERD and the risk of atrial fibrillation, and even the frequency of GERD was unrelated to the risk of atrial fibrillation. The results of this MR study align with Bunch et al. findings,[10] indicating that genetically predicted GERD is not associated with the risk of atrial fibrillation after eliminating the potential influence of confounding factors. Considering that the impact of confounding factors on the outcome was eliminated in this MR study, the conclusion is more accurate and reliable.
Additionally, this study has, for the first time, found that LDL cholesterol mediated 19.99% (95% CI: 4.49% to 35.50%), HDL cholesterol mediated 11.71% (95% CI: 5.23% to 18.19%), and hypertension mediated 35.09% (95% CI: 24.66% to 45.53%) of the effect of GERD on myocardial infarction, which has not been reported in previous research. This provides important evidence for reducing the risk of myocardial infarction in GERD patients, indicating that lowering LDL cholesterol, controlling hypertension, and increasing HDL cholesterol can help reduce the risk of myocardial infarction in GERD patients. Furthermore, this evidence potentially suggests that lipid-lowering and antihypertensive medications may significantly reduce the risk of myocardial infarction in GERD patients. However, this hypothesis still requires further evidence for clarification.
Potential mechanism
The observed causal relationship between GERD and alterations in blood pressure components, lipid profile, and increased risk of cardiovascular diseases suggests potential underlying mechanisms that warrant exploration. While the precise pathways remain to be fully elucidated, several hypotheses can be considered. GERD is known to trigger local and systemic inflammation.[38,39] Chronic inflammation caused by the presence of GERD may contribute to endothelial dysfunction,[40] atherosclerosis,[33] and subsequent cardiovascular events. Inflammatory mediators could potentially influence blood pressure regulation, lipid metabolism[41] and ultimately led to the occurrence of cardiovascular diseases. In addition, GERD-related esophageal irritation might damage cardiac autonomic dysfunction. Prior research suggests a disruption in the autonomic nervous system among individuals GERD, particularly affecting the sympathetic and parasympathetic components.[31] However, it seems that compromised parasympathetic nervous system function aligns more closely with the onset and development of GERD. Furthermore, GERD-associated alterations in lipid profile parameters could be linked to metabolic disturbances.[7,42] This may involve disruptions in lipid metabolism pathways, leading to increased levels of LDL cholesterol and triglycerides, and decreased levels of HDL cholesterol.
Clinical implication
The results of our MR study, elucidating the potential causal association between GERD and blood pressure components, lipid profile, and cardiovascular diseases, have substantial clinical implications. Firstly, healthcare providers should consider integrating cardiovascular risk assessments into the care plans for GERD patients. Regular monitoring of blood pressure components and lipid profiles may be necessary to identify individuals at a high risk of cardiovascular diseases. Secondly, the identified associations underscore the necessity for tailoring treatment strategies for GERD patients. Customizing interventions based on an individual’s cardiovascular risk profile, taking into account factors such as blood pressure and lipid profile, might improve overall treatment outcomes. Thirdly, with the potential causal association between GERD and cardiovascular diseases in mind, early intervention in GERD management may hold broader implications for cardiovascular health. Thus, addressing GERD comprehensively could play a role in mitigating cardiovascular risks. Finally, this MR study emphasizes the importance for research into therapeutic targets that simultaneously address both GERD symptoms and potential cardiovascular complications. Therefore, investigating medications or lifestyle interventions targeting both conditions could provide more holistic and effective treatment strategies.
Our study suggests that GERD is associated with alterations in blood pressure components, lipid profile parameters, and cardiovascular diseases, which could inform future guidelines and interventions aimed at improving patient outcomes. However, it is crucial to note that further research is needed to validate these findings and expand our understanding of the mechanisms underlying the observed relationships between GERD and cardiovascular/ metabolic complications. Firstly, longitudinal studies are necessary to further establish causal relationships between GERD and cardiovascular/metabolic outcomes in large sample size populations. Secondly, further research should focus on the biological mechanisms that link GERD to changes in blood pressure, lipid profiles, and cardiovascular diseases. This could involve exploring genetic, molecular, and physiological pathways.
Limitations
Several limitations need to be acknowledged. Firstly, we assume that our chosen genetic instruments adequately represent the entire spectrum of factors related to GERD. Considering the multifaceted nature of GERD with various environmental and genetic contributors, any gaps or inaccuracies in our representation may introduce bias into the estimates. Secondly, while MR is adept at establishing causal inference, it falls short in revealing the specific mechanisms underlying these associations. Further research is crucial to unravel the biological pathways linking GERD to blood pressure, lipid metabolism, and cardiovascular diseases. Thirdly, while we selected SNPs robustly associated with GERD from GWAS, there may still be unaccounted genetic variants influencing the outcomes through pathways other than GERD. Additionally, our findings may be affected by population characteristics of the GWAS data used. Lastly, our study concentrated on common genetic variants, potentially overlooking rare variants or gene-environment interactions that might influence the observed associations.
Conclusion
In summary, our MR study strongly suggests a potential cause and effect association between GERD and substantial alterations in blood pressure components, lipid profile, and an elevated risk of cardiovascular diseases. The identified links between genetically predicted GERD and increased SBP, DBP, MAP, LDL cholesterol, and triglycerides, along with a decreased level of HDL cholesterol, as well as a high risk of myocardial infarction and hypertension, highlight the complicated associations between GERD and cardiovascular health. Furthermore, this study has further discovered that LDL cholesterol, HDL cholesterol, and hypertension mediate the effect of GERD on myocardial infarction, providing important evidence for reducing the risk of myocardial infarction in GERD patients. These findings have significant clinical implications, emphasizing the need for cardiovascular risk assessment and personalized treatment strategies for individuals with GERD. However, further investigation is necessary to demonstrate the underlying mechanisms and confirm these associations across diverse populations.
Funding statement: None.
Acknowledgements
None.
-
Author Contributions
Q Wu, C He made equal contributions to this manuscript as co-first authors. D Lan, Q Zeng, and Q Su served as co-corresponding authors. Together, Q Wu, C He, D Lan, Q Zeng and Q Su took the lead in study design, data analysis, and drafting the original manuscript. C Song, W Huang, X Hao and Q Zeng were instrumental in shaping the study design, conducting the literature review, generating figures, and refining the manuscript through review and edits. Moreover, Q Wu, C He, C Song, X Hao, D Lan, Q Zeng, and Q Su were pivotal in conceptualizing the study, overseeing its execution, managing the project, and obtaining funding. W Huang and X Hao also played a crucial role by offering valuable advice and significantly enhancing the manuscript with their critical revisions for intellectual depth. All authors agree to the submission and publication of this study.
-
Ethical Approval
The summary-level data used were attained from publicly website, and ethical approval was not required as the data were de-identified and did not involve direct contact with study participants. This study adhered to ethical standards in the utilization of genetic and phenotypic data.
-
Informed Consent
The study design prioritized privacy and confidentiality, ensuring compliance with ethical guidelines governing the responsible use of genetic information. Additionally, transparency and adherence to established research ethics principles were maintained throughout the analysis and reporting process.
-
Conflict of Interest
The authors declare no competing interests.
-
Data Availability Statement
All the data used in this study can be freely downloaded from public databases (https://gwas.mrcieu.ac.uk/).
References
1 Maret-Ouda J, Markar SR, Lagergren J. Gastroesophageal reflux disease: a review. JAMA 2020;324:2536-2547.10.1001/jama.2020.21360Search in Google Scholar PubMed
2 Gyawali CP, Fass R. Management of gastroesophageal reflux disease. Gastroenterology 2018;154:302-318.10.1053/j.gastro.2017.07.049Search in Google Scholar PubMed
3 Richter JE, Rubenstein JH. Presentation and epidemiology of gastroesophageal reflux disease. Gastroenterology 2018;154:267-276.10.1053/j.gastro.2017.07.045Search in Google Scholar PubMed PubMed Central
4 Teragawa H, Oshita C, Ueda T. History of gastroesophageal reflux disease in patients with suspected coronary artery disease. Heart and Vessels. 2019;34:1631-1638.10.1007/s00380-019-01413-1Search in Google Scholar PubMed
5 Linz D, Hohl M, Vollmar J, Ukena C, Mahfoud F, Böhm M. Atrial fibrillation and gastroesophageal reflux disease: the cardiogastric interaction. Ep Europace 2017;19:16-20.10.1093/europace/euw092Search in Google Scholar PubMed
6 Ha JO, Lee TH, Lee CW, Park JY, Choi SH, Park HS, et al. Prevalence and risk factors of gastroesophageal reflux disease in patients with type 2 diabetes mellitus. Diabetes Metab J 2016;40:297-307.10.4093/dmj.2016.40.4.297Search in Google Scholar PubMed PubMed Central
7 Kallel L, Bibani N, Fekih M, Matri S, Karoui S, Mustapha N, et al. Metabolic syndrome is associated with gastroesophageal reflux disease based on a 24-hour ambulatory pH monitoring. Dis Esophagus 2011;24:153-159.10.1111/j.1442-2050.2010.01118.xSearch in Google Scholar PubMed
8 Huang CC, Chan WL, Luo JC, Chen YC, Chen TJ, Chung CM, et al. Gastroesophageal reflux disease and atrial fibrillation: a nationwide population-based study. PLoS One 2012;7:e47575.10.1371/journal.pone.0047575Search in Google Scholar PubMed PubMed Central
9 Maret-Ouda J, Santoni G, Xie S, Rosengren A, Lagergren J. Objectively confirmed gastroesophageal reflux disease and risk of atrial fibrillation: a population-based cohort study in Sweden. Eur J Gastroenterol Hepatol. 2022;34:1116-1120.10.1097/MEG.0000000000002419Search in Google Scholar PubMed
10 Bunch TJ, Packer DL, Jahangir A, Locke GR, Talley NJ, Gersh BJ, et al. Long-term risk of atrial fibrillation with symptomatic gastroesophageal reflux disease and esophagitis. Am J Cardiol 2008;102:1207-1211.10.1016/j.amjcard.2008.06.048Search in Google Scholar PubMed PubMed Central
11 VanderWeele TJ, Tchetgen EJT, Cornelis M, Kraft P. Methodological challenges in mendelian randomization. Epidemiology 2014;25:427.10.1097/EDE.0000000000000081Search in Google Scholar PubMed PubMed Central
12 Sanderson E, Glymour MM, Holmes MV, Kang H, Morrison J, Munafò MR, et al. Mendelian randomization. Nat Rev Methods Primers 2022;2:6.10.1038/s43586-021-00092-5Search in Google Scholar PubMed PubMed Central
13 Sekula P, Fabiola Del Greco M, Pattaro C, Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol 2016;27:3253.10.1681/ASN.2016010098Search in Google Scholar PubMed PubMed Central
14 Skrivankova VW, Richmond RC, Woolf BA, Davies NM, Swanson SA, VanderWeele TJ, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ 2021;375.10.1136/bmj.n2233Search in Google Scholar PubMed PubMed Central
15 Ong JS, An J, Han X, Law M, Nandakumar P, Me Research team, et al. Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett’s oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut 2022;71:1053-1061.10.1136/gutjnl-2020-323906Search in Google Scholar PubMed PubMed Central
16 Mbatchou J, Barnard L, Backman J, Marcketta A, Kosmicki JA, Ziyatdinov A, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet 2021;53:1097-1103.10.1038/s41588-021-00870-7Search in Google Scholar PubMed
17 Sakaue S, Kanai M, Tanigawa Y, Karjalainen J, Kurki M, Koshiba S, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet 2021;53:1415-1424.10.1038/s41588-021-00931-xSearch in Google Scholar PubMed
18 Klimentidis YC, Arora A, Newell M, Zhou J, Ordovas JM, Renquist BJ, et al. Phenotypic and genetic characterization of lower LDL cholesterol and increased type 2 diabetes risk in the UK Biobank. Diabetes 2020;69:2194-2205.10.2337/db19-1134Search in Google Scholar PubMed PubMed Central
19 Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Davey Smith G, et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med 2020;17:e1003062.10.1371/journal.pmed.1003062Search in Google Scholar PubMed PubMed Central
20 Shah S, Henry A, Roselli C, Lin H, Sveinbjörnsson G, Fatemifar G, et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 2020;11:163.10.1038/s41467-019-13690-5Search in Google Scholar PubMed PubMed Central
21 Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet 2018;50:1234-1239.10.1038/s41588-018-0171-3Search in Google Scholar PubMed PubMed Central
22 Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, et al. PhenoScanner: a database of human genotype–phenotype associations. Bioinformatics 2016;32:3207-3209.10.1093/bioinformatics/btw373Search in Google Scholar PubMed PubMed Central
23 Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133-1163.10.1002/sim.3034Search in Google Scholar PubMed
24 Mourtzi N, Georgakis MK, Ntanasi E, Hatzimanolis A, Ramirez A, Heilmann-Heimbach S, et al. Genetically downregulated Interleukin-6 signalling is associated with a lower risk of frailty. Age Ageing 2023;52:afac318.10.1093/ageing/afac318Search in Google Scholar PubMed
25 Ye Z, Zeng Q, Ning L, Huang W, Su Q. Systolic blood pressure is associated with abnormal alterations in brain cortical structure: Evidence from a Mendelian randomization study. Eur J Intern Med 2024;120:92-98.10.1016/j.ejim.2023.10.018Search in Google Scholar PubMed
26 Lin Z, Deng Y, Pan W. Combining the strengths of inverse-variance weighting and Egger regression in Mendelian randomization using a mixture of regressions model. PLoS Genet 2021;17:e1009922.10.1371/journal.pgen.1009922Search in Google Scholar PubMed PubMed Central
27 Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693-698.10.1038/s41588-018-0099-7Search in Google Scholar PubMed PubMed Central
28 Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304-314.10.1002/gepi.21965Search in Google Scholar PubMed PubMed Central
29 Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512-525.10.1093/ije/dyv080Search in Google Scholar PubMed PubMed Central
30 MacKinnon DP, Lockwood CM, Hoffman JM, West SG, Sheets V. A comparison of methods to test mediation and other intervening variable effects. Psychol Methods 2002;7:83.10.1037//1082-989X.7.1.83Search in Google Scholar
31 Milovanovic B, Filipovic B, Mutavdzin S, Zdravkovic M, Gligorijevic T, Paunovic J, et al. Cardiac autonomic dysfunction in patients with gastroesophageal reflux disease. World J Gastroenterol 2015;21:6982.10.3748/wjg.v21.i22.6982Search in Google Scholar PubMed PubMed Central
32 Kim YM, Kim Y, Kim J-H, Park JS, Baik SJ, Chun J, et al. Triglyceride-glucose index is associated with gastroesophageal reflux disease and erosive reflux disease: a health checkup cohort study. Sci Rep 2022;12:20959.10.1038/s41598-022-25536-0Search in Google Scholar PubMed PubMed Central
33 Lei WY, Wang JH, Wen SH, Yi CH, Hung JS, Liu TT, et al. Risk of acute myocardial infarction in patients with gastroesophageal reflux disease: A nationwide population-based study. PLoS One 2017;12:e0173899.10.1371/journal.pone.0173899Search in Google Scholar PubMed PubMed Central
34 Eisa M, Sandhu A, Prakash R, Ganocy SJ, Fass R. The risk of acute myocardial infarction in patients with gastroesophageal reflux disease. J Neurogastroenterol Motil 2020;26:471.10.5056/jnm19192Search in Google Scholar PubMed PubMed Central
35 Li ZT, Ji F, Han XW, Wang L, Yue YQ, Wang ZG. The role of gastroesophageal reflux in provoking high blood pressure episodes in patients with hypertension. J Clin Gastroenterol 2018;52:685.10.1097/MCG.0000000000000933Search in Google Scholar PubMed PubMed Central
36 He SY, Liu Y, Xu J, Luo G, Cao L, Long X. Prevalence and predictors of silent gastroesophageal reflux disease in patients with hypertension. Gastroenterol Res Pract 2018;2018:7242917.10.1155/2018/7242917Search in Google Scholar PubMed PubMed Central
37 Sun X, Chen L, Zheng L. A Mendelian randomization study to assess the genetic liability of gastroesophageal reflux disease for cardiovascular diseases and risk factors. Hum Mol Genet 2022;31:4275-4285.10.1093/hmg/ddac162Search in Google Scholar PubMed
38 Rieder F, Biancani P, Harnett K, Yerian L, Falk GW. Inflammatory mediators in gastroesophageal reflux disease: impact on esophageal motility, fibrosis, and carcinogenesis. Am J Physiol Gastrointest Liver Physiol 2010;298:G571-G81.10.1152/ajpgi.00454.2009Search in Google Scholar PubMed PubMed Central
39 Yoshida N. Inflammation and oxidative stress in gastroesophageal reflux disease. J Clin Biochem Nutr 2007;40:13-23.10.3164/jcbn.40.13Search in Google Scholar PubMed PubMed Central
40 Kandulski A, Malfertheiner P. Gastroesophageal reflux disease—from reflux episodes to mucosal inflammation. Nat Rev Gastroenterol Hepatol 2012;9:15-22.10.1038/nrgastro.2011.210Search in Google Scholar PubMed
41 Chang ML, Yang Z, Yang SS. Roles of adipokines in digestive diseases: markers of inflammation, metabolic alteration and disease progression. Int J Mol Sci 2020;21:8308.10.3390/ijms21218308Search in Google Scholar PubMed PubMed Central
42 Lee YC, Yen AMF, Tai JJ, Chang SH, Lin JT, Chiu HM, et al. The effect of metabolic risk factors on the natural course of gastro-oesophageal reflux disease. Gut 2009;58:174-181.10.1136/gut.2008.162305Search in Google Scholar PubMed PubMed Central
© 2024 Qiang Wu, Changjing He, Wanzhong Huang, Chaoqun Song, Xin Hao, Qing Zeng, Dazhi Lan, Qiang Su, published by De Gruyter on behalf of the SMP
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Review Article
- Histological transformation in lung adenocarcinoma: Insights of mechanisms and therapeutic windows
- Original Article
- Inetetamab for injection in combination with vinorelbine weekly or every three weeks in HER2-positive metastatic breast cancer: A multicenter, randomized, phase II clinical trial
- Exosomes derived from Umbilical cord mesenchymal stem cell promote hair regrowth in C57BL6 mice through upregulation of the RAS/ERK signaling pathway
- Integrated untargeted/targeted metabolomics identifies a putative oxylipin signature in patients with atrial fibrillation and coronary heart disease
- Gastroesophageal reflux disease influences blood pressure components, lipid profile and cardiovascular diseases: Evidence from a Mendelian randomization study
- Rapid Communication
- Prognostic prediction of m6A and ferroptosis-associated lncRNAs in liver hepatocellular carcinoma
- Erratum
- Erratum to "WWP2 protects against sepsis-induced cardiac injury through inhibiting cardiomyocyte ferroptosis"
Articles in the same Issue
- Review Article
- Histological transformation in lung adenocarcinoma: Insights of mechanisms and therapeutic windows
- Original Article
- Inetetamab for injection in combination with vinorelbine weekly or every three weeks in HER2-positive metastatic breast cancer: A multicenter, randomized, phase II clinical trial
- Exosomes derived from Umbilical cord mesenchymal stem cell promote hair regrowth in C57BL6 mice through upregulation of the RAS/ERK signaling pathway
- Integrated untargeted/targeted metabolomics identifies a putative oxylipin signature in patients with atrial fibrillation and coronary heart disease
- Gastroesophageal reflux disease influences blood pressure components, lipid profile and cardiovascular diseases: Evidence from a Mendelian randomization study
- Rapid Communication
- Prognostic prediction of m6A and ferroptosis-associated lncRNAs in liver hepatocellular carcinoma
- Erratum
- Erratum to "WWP2 protects against sepsis-induced cardiac injury through inhibiting cardiomyocyte ferroptosis"

