Abstract
Objectives
Colorectal cancer (CRC) is one of the most prevalent cancer types worldwide, exhibiting significant variance in incidence rates across different ethnicities and geographical regions. Notably, there is a rising incidence of CRC among younger adults, particularly evident in advanced stages, with a more pronounced trend observed in developing nations. Epigenetic alterations potentially play a role in the early onset of CRC and could elucidate interpopulation disparities. This study aimed to examine DNA methylation levels in the tumor suppressor genes MLH1 and p16INK4a, comparing Nepalese and Swedish patients with CRC.
Methods
Patients who underwent CRC surgery at Tribhuvan University Teaching Hospital, Nepal (n=39), and Sahlgrenska University Hospital, Sweden (n=39) were included. Demographic and clinicopathological data were analyzed, and pyrosequencing was employed to determine methylation levels in the MLH1 promoter region and the first exon of p16INK4a in tumor tissues and adjacent mucosa located 10 cm from the tumor site. Subsequently, methylation status was compared between Nepalese and Swedish patients and correlated with clinicopathological parameters.
Results
Nepalese and Swedish patients displayed equal levels of MLH1 and p16INK4a methylation in tumors, but Nepalese patients exhibited a significantly higher level of MLH1 methylation in mucosa compared to Swedish patients (p=0.0008). Moreover, a greater proportion of Nepalese patients showed MLH1 methylation in mucosa compared to Swedish patients (31 vs. 2.6 %). Aberrant methylation of p16INK4a was also observed in the mucosa of Nepalese patients, characterized by high methylation at specific sites rather than uniform methylation across CpG sites. There were no significant differences in methylation levels based on tumor location among Nepalese patients, whereas Swedish patients exhibited higher methylation in right- compared to left-sided colon tumors. Swedish patients showed an increase in p16INK4a methylation in tumors with advancing age.
Conclusions
Nepalese and Swedish patients displayed equal levels of MLH1 and p16INK4a methylation in tumors. In contrast, Nepalese patients had a higher level of MLH1 methylation as well as aberrant methylation of p16INK4a in mucosa compared to Swedish patients. These epigenetic differences may be linked to environmental and lifestyle factors. Ongoing research will further explore whether hypermethylation in the mucosa of Nepalese patients is associated with tumorigenesis and its potential utility in screening high-risk patients or predicting recurrence.
Introduction
Colorectal cancer (CRC) ranks as the third most prevalent cancer disease worldwide, yet its incidence rates exhibit a staggering tenfold variation globally. Notably, European societies report the highest rates, ranging between 40 and 90 cases per 100,000 individuals, while many African regions, like middle Africa, demonstrate significantly lower rates, at approximately 12 cases per 100,000 [1]. Similarly, South Asia presents a diverse landscape, with affluent societies such as South Korea, Japan, and Singapore reporting higher rates compared to lower-income countries in the region [2]. In India, the age-adjusted incidence rate, standardized against a global population standard, drops to as low as 4 cases per 100,000 individuals [2].
The primary risk factor attributed to the onset of CRC is advancing age [3]. European studies estimate a mean age of 72 years among patients with CRC, while in low-income Asian countries, this figure ranges from 43.7 to 51.2 years [4, 5]. Disparities in age between affluent and low-income nations may significantly contribute to the notable divergence in CRC incidence rates within these populations. Interestingly, while there has not been a general increase in CRC incidence over recent decades, there has been a distinct rise observed among individuals under 50 years of age [6]. Additionally, a decline in the age of CRC diagnosis is evident in Asian countries [7]. A retrospective analysis spanning a decade (1999–2008) at a tertiary hospital in Nepal uncovered a noteworthy trend: the mean age of patients with CRC decreased from 34±4.7 years in the initial 5 years to 31±5.1 years in the subsequent half, indicating a declining trend in the age at diagnosis [8].
Numerous studies have delved into the genetic and epigenetic changes underlying the onset of CRC. Of particular interest is DNA hypermethylation, a crucial process implicated in the suppression of tumor suppressor genes. This aberrant methylation of CpG-rich promoter regions of tumor suppressor genes leads to transcriptional inactivation, a hallmark of CRC development [9]. These alterations manifest early and have been discerned in macroscopically normal mucosa adjacent to both benign and malignant tumors. Primarily, they occur at cytosine bases within CpG dinucleotides outside CpG islands [10]. Additionally, DNA replication errors in microsatellite repeat sequences can result in microsatellite instability (MSI), observed in 10–15 % of sporadic CRC cases, signaling deficiencies in DNA mismatch repair (MMR) genes. Epigenetic modifications in the MutL homolog 1 gene (MLH1) frequently contribute to MSI, particularly through alterations in its promoter region, which is widely recognized as an early event in CRC tumorigenesis [11], [12], [13].
Frequent hypermethylation has also been documented in the INK4b-ARF-INK4a locus, which encodes two critical cyclin-dependent kinase inhibitors, p15INK4a and p16INK4a, both pivotal in regulating the G1 phase of the cell cycle. Within this locus, an additional protein, p14ARF (alternative reading frame), is also encoded [14]. A prior investigation revealed that hypermethylation of the p16INK4a gene promoter was detected in 36 % of mucosa samples obtained 10 cm from the tumor in patients with CRC [15]. Notably, hypermethylation of p16INK4a in mucosa correlated with poorer patient outcomes, suggesting that epigenetic changes in the surrounding mucosa might precede genetic alterations in the early stages of carcinogenesis [13].
While epigenetics has been extensively studied in Western populations, the association between epigenetics and CRC remains largely unexplored in resource-constrained countries such as Nepal. This study aimed to bridge this gap by comparing DNA methylation levels in two commonly hypermethylated tumor suppressor genes, MLH1 and p16INK4a, within the tumor and matched mucosa of patients with CRC from both Nepal and Sweden. Additionally, the study aimed to analyze the correlation between the methylation data and the demographic and clinicopathological profiles of the patients.
Patients and methods
Patients
Nepalese patients (n=39) who underwent surgery for sporadic (nonhereditary) CRC at Tribhuvan University Teaching Hospital during 2013–2015, and from whom there were accessible tissues and clinical data, were consecutively enrolled without any bias in patient selection. These Nepalese patients were matched for age and sex with 39 Swedish patients who underwent CRC surgery at Sahlgrenska University Hospital. Demographic and clinicopathological data, including age, sex, tumor site, tumor stage, tumor grade, histopathological type, and lymph node status, were recorded. The tumor location was classified as the right colon (cecum, ascending colon, hepatic flexure, and proximal third of the transverse colon), left colon (distal transverse colon, splenic flexure, descending colon, and sigmoid colon), and rectum [16].
Tissue sampling
Tissue samples were collected during surgery from both tumor sites and macroscopically normal-appearing mucosa situated 10 cm away from the tumor. Tissue samples from Swedish patients were snap-frozen in liquid nitrogen and stored at −80 °C until analysis. In contrast, tissues from Nepalese patients were formalin-fixed and paraffin-embedded (FFPE). To ensure the reliability of the results, methylation levels were assessed and compared in five matched fresh-frozen and FFPE tumor tissues (Supplementary Table 1). Hematoxylin and eosin (H&E) staining was employed for histological confirmation of both FFPE tumor and mucosa tissues.
DNA extraction
Genomic DNA was isolated from frozen tissue using Qiagen AllPrep DNA/RNA/Protein Kit and from 10 μm tissue sections using the Qiagen FFPE All prep DNA/RNA Kit, according to the manufacturer’s instructions. The DNA samples were kept at −20 °C until analysis.
DNA denaturation and bisulfite conversion
Two hundred ng of genomic DNA from each sample were used for bisulfite conversion and bisulfite treatment was performed using EpiTecht® Fast Bisulfite Kit (Qiagen, Sollentuna, Sweden) according to the manufacturer’s protocol.
Measurement of methylation levels
The methylation levels of the MLH1 and p16INK4a genes were quantified using pyrosequencing and PyroMark Q24 assays (Qiagen, Sollentuna, Sweden). The MLH1 sequence to analyze included five CpG sites: YGGATAGYGATTTTTAAYGYGTAAGYGTATA.
The p16INK4a sequence to analyze included six CpG sites: TYGTTAAGTGTTYGGAGTTAATAGTATTTTTTTYGAGTATTYGTTTAYGGYGT; however, only five CpG sites were included in the analyses due to the presence of a single nucleotide polymorphism at the fifth CpG site position. The polymerase chain reactions (PCR) were performed according to the manufacturer’s instructions.
Preparation of samples for pyrosequencing
One μL of Sepharose beads were mixed with 40 μL of binding buffer and 29 μL of water in an Eppendorf tube. About 60 μL of this mix were added to 20 μL of PCR products in a 96 well plate and agitated at 1,500 rpm for 10 min. The sequencing primers were diluted to 0.375 μM with PyroMark annealing buffer and 20 μL of this solution were then transferred to the PyroMark Advanced Q24 Plate. The washes were performed using the vacuum station according to the manufacturer’s instruction. To anneal the samples to sequencing primers, the temperature was increased to 80 °C for 5 min and the plates were then immediately transferred to the PyroMark Advanced Q24 instrument for processing.
Pyrosequencing
Pyrosequencing of the purified single stranded PCR products and CpG site quantification was accomplished by the PyroMark Q24 and related software (Qiagen). The CpG sites were investigated in both mucosa and tumor samples. Each CpG site was assigned a percentage of methylation by evaluating the C/T ratio. The mean percentage of methylation across the CpG sites was obtained for each analyzed gene. Representative pyrograms are presented in Supplementary Figures 1 and 2. Hypermethylation was defined as a mean methylation value above the highest value found at any CpG site in normal mucosa obtained from controls in a previous study. This value was 5 % for the MLH1 gene and 6 % for the p16INK4a gene (data not shown).
Statistical analysis
The obtained data were analyzed by statistical modeling using the commercial software JMP 15.0.0 (SAS Institute, 2019). Data are presented as mean values with SEM bars or as medians and ranges. Differences between groups were assessed using the nonparametric Wilcoxon/Kruskal–Wallis test. p-values <0.05 were considered significant. No correction for multiple testing was done.
Results
In the study, 56 % of participants were male, and 44 % were female. The median age was 53 years, ranging from 19 to 78 years. Stage III CRC was predominant in the Nepalese group (54 %) compared to the Swedish group (38 %). The majority of the Nepalese patients exhibited well-differentiated tumors (41 %), whereas moderately differentiated tumors were more common among Swedish patients (53 %). Colon cancer represented 74 % of cases among Nepalese patients and 62 % among Swedish patients, with the predominant site of lesions being the right colon (Table 1).
Patient and tumor characteristics.
| Nepalese patients | Swedish patients | |
|---|---|---|
| Median age, years (range) | 52 (20–78) | 53 (19–78) |
|
|
||
| Sex, n, % | ||
|
|
||
| Male | 22 (56.4) | 22 (56.4) |
| Female | 17 (43.6) | 17 (43.6) |
|
|
||
| Tumor stage, n, % | ||
|
|
||
| I | 3 (8.6) | 6 (15.4) |
| II | 13 (37.1) | 11 (28.2) |
| III | 19 (54.3) | 15 (38.5) |
| IV | 0 | 7 (17.9) |
|
|
||
| Tumor differentiation, n, % | ||
|
|
||
| Well | 14 (41.2) | 2 (5.3) |
| Moderate | 13 (38.2) | 20 (52.6) |
| Low | 7 (20.6) | 11 (28.9) |
| Mucinous | 0 | 5 (13.2) |
|
|
||
| Tumor location, n, % | ||
|
|
||
| Right side of colon | 18 (47.4) | 17 (43.6) |
| Left side of colon | 10 (26.3) | 7 (17.9) |
| Rectum | 10 (26.3) | 15 (38.5) |
MLH1 methylation at individual CpG sites
MLH1 methylation levels could be determined in all samples from both the Nepalese and Swedish patient cohorts. Distinct variations in MLH1 methylation levels between the mucosa and tumors of cohorts were evident at individual CpG sites (Figure 1, Supplementary Table 2). In mucosa samples from Nepalese patients, MLH1 methylation levels ranged from 0 to 50 %, contrasting with 0 to 8% in Swedish patients. MLH1 methylation levels in tumor samples ranged from 0 to 46 % and 0 to 74 % in the two patient cohorts, respectively.
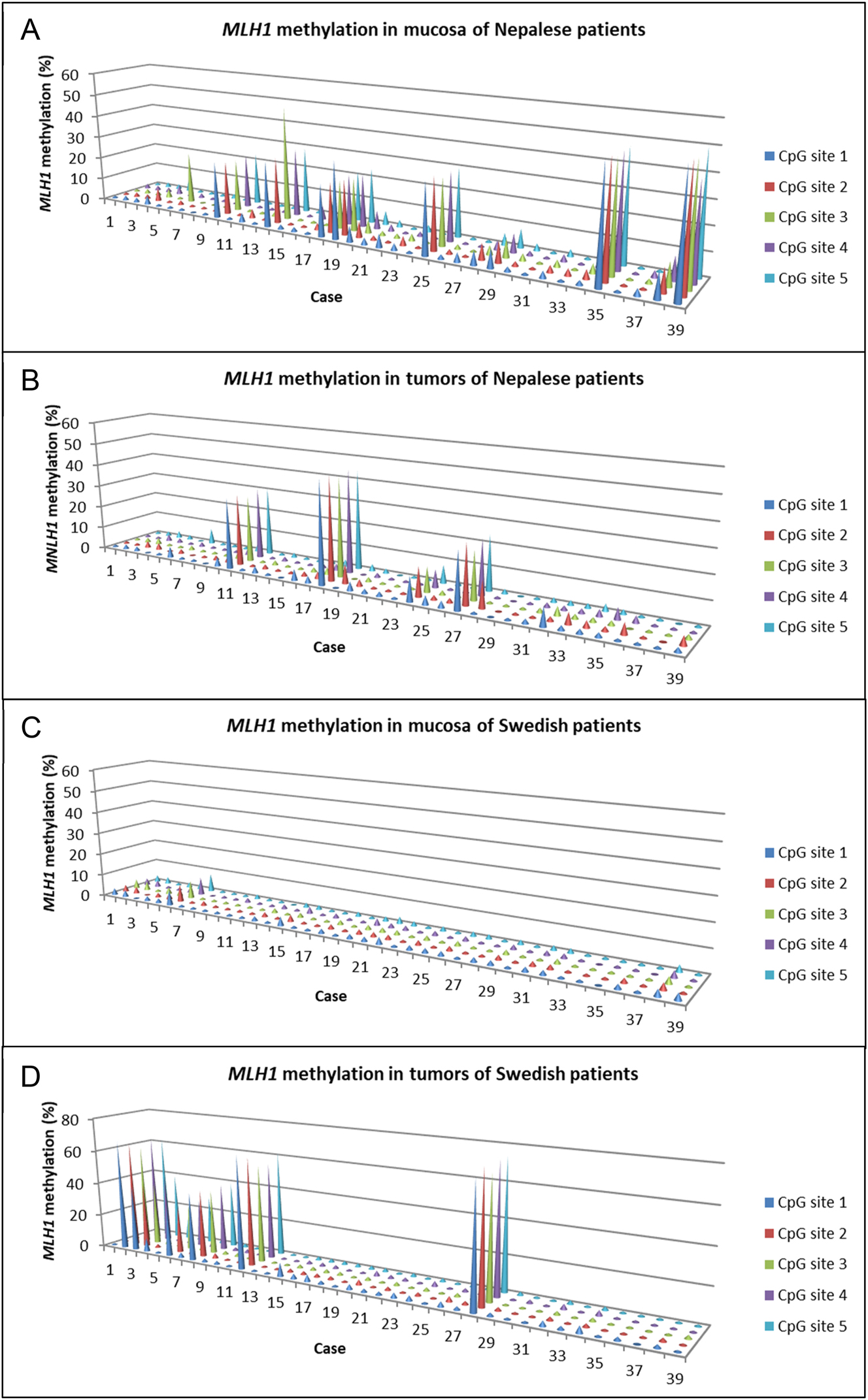
MLH1 methylation at each CpG site in mucosa and tumor tissue of Nepalese patients (A and B) and Swedish patients (C and D) with colorectal cancer.
p16INK4a methylation at individual CpG sites
p16INK4a methylation was assessable in all samples from Swedish patients and in 32 out of 39 mucosa samples and 31 out of 39 tumor samples from Nepalese patients. In contrast to the generally uniform methylation pattern observed for MLH1 in mucosa, there was notable variability in methylation levels among different CpG sites for p16INK4a, particularly with elevated levels at CpG sites 4 and 6 in mucosa samples from Nepalese patients (Figure 2). Methylation levels of p16INK4a ranged from 0 to 46 % in mucosa samples from Nepalese patients, contrasting with 0 to 21 % in Swedish patients. Although p16INK4a hypermethylation was more prevalent in tumor samples from Swedish patients, the highest methylation level was detected in a tumor from one of the Nepalese patients (case 19, Figure 3B). Methylation levels of p16INK4a in tumors ranged from 1 to 100 % and from 1 to 75 % in the Nepalese and Swedish patient cohorts, respectively.

p16INK4a methylation at each CpG site in mucosa and tumor tissue of Nepalese patients (A and B) and Swedish patients (C and D) with colorectal cancer. CpG site 5 was not included due to a single nucleotide polymorphism at this position.
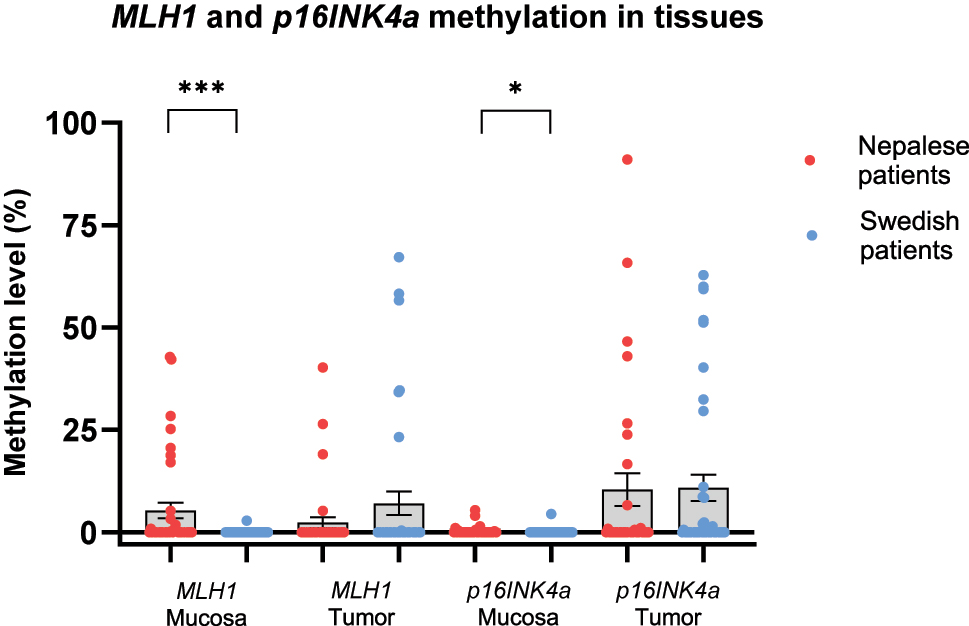
Scatter dot plots showing MLH1 and p16INK4a methylation levels in mucosa and tumor tissues of (A) Nepalese and (B) Swedish patients with colorectal cancer. Data are presented as mean values with SEM bars.
MLH1 and p16INK4a methylation in mucosa and tumor tissue
The mean methylation levels of MLH1 and p16INK4a in mucosa and tumors are illustrated in Figure 3. As shown, the methylation levels of MLH1 and p16INK4a were significantly higher in mucosa of Nepalese, compared to Swedish patients. It should be noted though, that the elevated methylation levels of p16INK4a in mucosa of Nepalese patients primarily originated from high methylation at CpG site 4, as depicted in Figure 2. Upon excluding this site from the mean value calculations, the difference between the two groups ceased to be statistically significant. There was no significant difference in MLH1 or p16INK4a methylation levels within tumors between the two patient cohorts.
Methylation according to age
Patients were stratified into three age groups (<50, 50–60, and >60 years old) and assessed for MLH1 and p16INK4a methylation in both mucosa and tumor tissue. The results are presented in Figure 4. There was no significant difference in methylation levels between age groups within the Nepalese patient cohort. However, Swedish patients aged over 60 years exhibited a higher p16INK4a methylation level compared to those younger than 50 years (p=0.0046). Furthermore, Nepalese patients had a higher MLH1 methylation level in mucosa compared to Swedish patients in the age groups of 50–60 (p=0.011) and >60 (p=0.016). The two Swedish patients displaying either MLH1 or p16INK4a methylation in mucosa belonged to the youngest age group. Additional details regarding the percentages of Nepalese and Swedish patients with MLH1 hypermethylation at respective CpG sites in mucosa and tumor tissue, segmented by age group, can be found in Supplementary Table 3.

Scatter dot plots showing MLH1 and p16INK4a methylation levels in mucosa and tumor tissues of (A) Nepalese and (B) Swedish patients with colorectal cancer by age group. Data are presented as mean values with SEM bars.
Among Swedish patients, the percentage of individuals lacking both MLH1 and p16INK4a methylation in tumor samples decreased with age, dropping from 82 % in the <50 years age group to 33 % in the >60 years age group (Figure 5A). Conversely, in the Nepalese patient cohort, as much as 75 % of patients in the >60 years age group lacked methylation of both genes.

Bars and boxes comparing the MLH1 and p16INK4a methylation status in tumors deriving from Nepalese and Swedish patients with colorectal cancer by (A) age group and (B) sex. The methylation status is classified as follows: no methylation, exclusive MLH1 methylation, exclusive p16INK4a methylation, or methylation in both genes.
Methylation according to sex
A notable percentage of Nepalese patients exhibited MLH1 methylation in mucosa without concurrent methylation in tumor tissue, irrespective of gender, whereas the single Swedish patient (female) with MLH1 methylation in mucosa also displayed methylation in tumor (Supplementary Figure 3A). Conversely, the proportion of Nepalese patients with methylation exclusively in tumor tissue was low among both females and males (5.9 vs. 9.1 %). In contrast, a higher percentage of Swedish female patients (29.4 %) showed MLH1 methylation exclusively in tumor tissue compared to males (4.5 %).
Exclusive p16INK4a methylation in mucosa without concurrent methylation in tumor tissue was solely detected in female Nepalese patients (Supplementary Figure 3B). However, some patients, both female and male, exhibited methylation in both mucosa and tumor. Similarly, a subset of both female and male patients within the Nepalese cohort displayed exclusive p16INK4a methylation within tumor tissue. Conversely, in the Swedish cohort, almost twice as many females as male patients displayed p16INK4a methylation exclusively in tumor tissue; however, one male patient exhibited p16INK4a methylation in both tumor and mucosa. Upon analyzing the sex distribution based on the presence of both MLH1 and p16INK4a hypermethylation in tumor tissue, it was more prevalent among females than males in both patient cohorts (Figure 5B). The highest frequency was observed among Swedish females (29.4 %).
Methylation according to tumor location
No statistically significant difference was observed in the methylation levels of MLH1 or p16INK4a in mucosa or tumor tissues among Nepalese patients based on tumor location (Figure 6A). In Swedish patients, however, significantly higher methylation levels of both genes were detected in tumor samples obtained from the right side of the colon (Figure 6B).
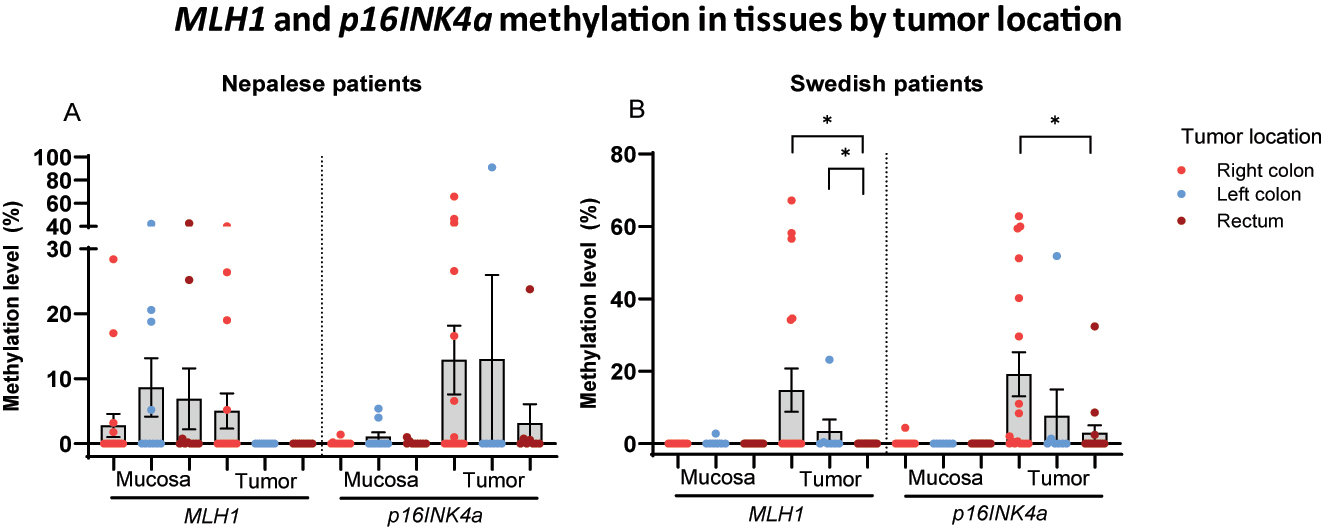
Scatter dot plots showing MLH1 and p16INK4a methylation levels in mucosa and tumor tissues of (A) Nepalese and (B) Swedish patients with colorectal cancer by tumor site. Data are presented as mean values with SEM bars.
Methylation according to tumor stage and differentiation
No significant differences in methylation levels were observed across tumor stages, except for MLH1 methylation in stage III tumors of Swedish patients, which exhibited higher levels compared to other stages (p=0.049). Furthermore, no significant differences in methylation levels were noted based on tumor differentiation.
Discussion
In this study, we compared the methylation status of the tumor suppressor genes MLH1 and p16INK4a in colorectal tumor tissue and matching mucosa samples from Nepalese and Swedish patients with CRC. Our findings revealed a significantly higher frequency of DNA methylation in the mucosa of Nepalese patients compared to Swedish patients. Additionally, the methylation levels observed in some mucosa samples from Nepalese patients were notably elevated compared to previous reports [17]. The underlying reason for this unexpectedly high methylation level in the mucosa of Nepalese patients remains unclear, but it may be linked to environmental and lifestyle factors known to influence gene regulation through epigenetic mechanisms, including DNA methylation. Such factors encompass age, sex, diet, lifestyle choices, environmental exposures, and medications [18, 19] as discussed below.
Age-related promoter hypermethylation has been documented for several tumor suppressor genes, including the mismatch repair gene MLH1, which contributes to 15–30 % of stomach and colon carcinomas [20]. Additionally, it is known that hypermethylation is more prevalent among females than males. Consequently, age and sex differences between patient groups may yield disparate results. However, in this study, the two patient cohorts were matched for these covariates; thus, any observed differences in methylation status between groups would likely be attributed to other factors [18]. Nevertheless, MLH1 hypermethylation at all five analyzed CpG sites in DNA derived from mucosa was more common in older Nepalese patients. Moreover, the percentage of patients with methylation of both MLH1 and p16INK4a in tumors was observed to increase with age in both Nepalese and Swedish patients. Consistent with previous reports, both MLH1 and p16INK4a methylation in tumor tissue was more prevalent in females compared to males among Swedish patients [21, 22]. MLH1 methylation in the mucosa of Nepalese patients was also more prevalent in females. However, the methylation status in tumors of Nepalese patients did not follow this pattern, as cases with MLH1 and p16INK4a hypermethylation in tumors were equally distributed among female and male patients.
In addition to age and sex, the occurrence of hypermethylation may also be influenced by tumor location. Typically, hypermethylation is more prevalent in tumors situated on the right side compared to those on the left side of the colon [23]. Consistent with previous findings, the results of this study revealed a higher level of methylation in right-sided colon tumors among both Nepalese and Swedish patients. However, methylation levels in mucosa samples did not exhibit variations based on tumor location.
There was no observed correlation between the methylation levels in mucosa and tumor samples within any of the patient cohorts. This finding aligns with similar results reported by Coppede et al., who analyzed five CRC-related genes in patients with CRC [19]. Among these genes, APC exhibited the highest frequency of methylation in both mucosa and tumor tissue, yet the methylation level in mucosa remained below 10 % and did not correlate with that in tumors. Conversely, Ramirez et al., in a methylation analysis of six genes including MLH1 and p16INK4a, found that up to 71 % of patients exhibited similar methylation patterns between mucosa and tumor tissues [13]. Additionally, 47 % of the mucosa samples displayed methylation in one or two loci, while 12 % showed methylation in three or more loci. The concept of “field cancerization” has been proposed in normal mucosa adjacent to tumor cells [24]. The presence of hypermethylation in mucosa cells of patients with colon cancer indicates that these cells harbor some of the epigenetic alterations associated with the tumor [25]. Such alterations in the mucosa surrounding CRC lesions may serve as predictors of outcome. As demonstrated in a previous study, hypermethylation of p16INK4a in CRC mucosa was associated with a worse disease-free survival [26].
The incidence of CRC is progressively rising, particularly in low- and middle-income countries, likely due to shifting demographics and dietary patterns [27]. Notably, Asians relocating to the United States eventually develop a similar CRC incidence as native-born Americans [28], suggesting a potential influence of dietary habits. It has been posited that the prevalent vegetarian diet in Asian and African societies may confer protection against CRC [29]. Obesity, strongly linked to CRC, is also recognized as a risk factor for DNA hypermethylation [30, 31]. In Sweden, the prevalence of obesity has been steadily increasing over recent decades, whereas Nepalese patients may experience undernourishment comparatively [32]. Obesity could thus contribute to a higher incidence of DNA hypermethylation and CRC development in Swedish patients compared to Nepalese patients. Moreover, dietary differences may play another significant role: the predominant Nepali vegetarian diet may shape a gut microbiota distinct from that resulting from a typical Western-style diet prevalent in Sweden. Additionally, environmental factors vary between the two populations. For instance, Kathmandu, the capital of Nepal, faces significant pollution, and its tap water is often contaminated with bacteria and parasites like Giardia Lamblia. These diverse factors, known to influence DNA methylation and subsequent gene regulation and expression, underscore the disparities between the Nepalese and Swedish populations [18].
Currently, we can only speculate on the reasons for the high methylation levels found in the mucosa of Nepalese patients. One hypothesis would be that aberrant methylation may be triggered in large areas of the gut in response to alterations in the microbiota. The interesting connection between the microbiota and the onset of CRC is an emerging area of research that is garnering increasing attention. For example, Fusobacterium nucleatum has been linked to promoter methylation of MLH1 and p16INK4a, while Streptococcus spp. seem to be associated with MLH1 hypermethylation [33]. In another recent study involving mice, even a nonpathogenic microbiota was found to significantly influence DNA methylation in the gut, contributing to the development of aberrant methylomes observed in aging and cancer [34].
With the increasing incidence of CRC among individuals under 50 years old, there is a recent recommendation to lower the age for CRC screening to 45 years [35]. However, routine colonoscopy screening may not be feasible in all countries, particularly those with low CRC incidence. Nevertheless, analyzing methylation in mucosa samples obtained during colonoscopy could serve as a screening tool for high-risk patients and potentially as a follow-up tool for predicting recurrent disease. Moreover, ongoing research is focused on developing and investigating novel techniques for detecting and quantifying methylated DNA using less invasive methods.
The carcinoembryonic antigen (CEA) marker is utilized to monitor treatment response in patients with CRC, while the Fecal Occult Blood Test (FOBT) has been employed for screening high-risk populations; however, its sensitivity and specificity, particularly for proximal CRC, are limited [36]. Given this context, detecting epigenetic alterations in body fluids could offer a safer and more effective screening method for CRC in high-risk populations [27]. Recently, the U.S. Food and Drug Administration (FDA) approved methylated Septin 9 (SEPT9) as a diagnostic marker for CRC screening. SEPT9 exhibits higher sensitivity than the fecal immunochemical test (FIT) and CEA in CRC detection [37]. Nevertheless, SEPT9 analysis is blood-based, and utilizing methylation tests based on fecal samples could provide an effective noninvasive alternative for diagnosing, screening, and monitoring CRC. However, CpG island methylation of certain tumor suppressor genes, including MLH1 and p16INK4a, can also be detected in both mucosal and fecal DNA of healthy individuals. Therefore, it is crucial to identify informative CRC-associated markers whose levels significantly differ from the normal baseline [17].
Strengths and limitations of study
Extensive research on CRC epigenetics has been conducted in Western populations, but little is known about this phenomenon in resource-constrained countries like Nepal. To our knowledge, this study marks the first investigation of tumor suppressor gene methylation in tumor and mucosa tissue among a Nepalese population with CRC. However, the study is subject to limitations, primarily the relatively small number of subjects studied. This limitation stemmed mainly from the low incidence of CRC in Nepal. In developing countries such as Nepal and India, a considerable proportion of patients with CRC exhibit metastasis at diagnosis, and a significant percentage undergo palliative procedures, precluding the collection of tissue from mucosa and tumors [8, 38]. Nevertheless, the study groups were matched for age and sex, which is a strength, given that these covariates are often associated with DNA methylation status.
There was a concern that the results might have been biased due to the use of different tissue preservation methods in the two patient cohorts. However, upon comparing methylation data from matched FFPE and fresh-frozen tumors, a high degree of similarity was observed. Moreover, Oliveira et al. [39] investigated the performance of DNA methylation analysis in FFPE vs. fresh-frozen ovarian cancer samples and found overall DNA methylation profiles to exhibit high concordance. Additionally, Moran et al. [40] generated single-base resolution DNA methylation profiles covering approximately 5.5 million CpG sites across the genome. Profiles from matched FFPE and fresh-frozen samples were compared to assess the impact of FFPE-related artifacts on methylation calls, and no significant inadequacies were identified. p16INK4a methylation status was unavailable for eight mucosa and tumor samples from the Nepalese patient cohort. The negative pyrosequencing results may have been due to formalin-induced DNA degradation, affecting bisulfite conversion and PCR amplification of the DNA. However, DNA of sufficient quality could be extracted from the remaining FFPE samples, as indicated by the internal control assessing successful bisulfite treatment and the high CpG peaks in the pyrograms.
Conclusions
Nepalese and Swedish patients displayed equal levels of MLH1 and p16INK4a methylation in tumors, but Nepalese patients had a higher level of MLH1 and p16INK4a methylation in mucosa compared to Swedish patients. These discrepancies may be attributed to environmental and lifestyle factors known to influence gene regulation through epigenetic mechanisms, including DNA methylation. Whether the elevated hypermethylation level in the mucosa of Nepalese patients with CRC represents an early aberration during tumorigenesis that could be valuable for screening high-risk patients or predicting recurrence is currently unknown. Ongoing studies will address this question by assessing the prevalence of tumor suppressor gene hypermethylation in the colorectal mucosa of Nepalese control subjects.
Funding source: The University of Gothenburg via the Global University Initiative
Funding source: The Swedish state under the LUA-ALF agreement
Award Identifier / Grant number: ALFGBG-542821
Funding source: The Ingabritt and Arne Lundberg Foundation
Award Identifier / Grant number: 335/07
Acknowledgments
We would like to thank S. Shrestha at the Pathology Department and Dr. A. Bhattarai at Biochemistry Department for help with analysis of data in Nepal. We acknowledge A. Helminen, H. Björkquist, and L. Munro for collecting biopsy samples and for work with the clinical database in Sweden. We are grateful to J. Flach and M. Åkerström for skillful technical assistance.
-
Research ethics: The study was approved by the Regional Ethical Review Boards in Kathmandu (ethical board no. 854/2019) and Gothenburg (ethical board no. 118–15), respectively, and was conducted in accordance with the Declaration of Helsinki.
-
Informed consent: Informed consent was obtained from all individuals included in this study.
-
Author contributions: Y.W., B.G., and G.K. designed the study. B.G. and Y.S. recruited the Nepalese study subjects and collected samples at surgery. P.F. prepared tissue sections for DNA extraction. Y.W. developed the methodology and performed methylation analysis. B.G. and Y.W. analyzed the data and performed statistics. All authors wrote, read, revised, and approved the final manuscript.
-
Competing interests: The authors state no conflict of interest.
-
Research funding: This work was supported by grants from the Swedish state under the LUA-ALF agreement [ALFGBG-542821], the University of Gothenburg via the Global University initiative, and the Ingabritt and Arne Lundberg Foundation [335/07]. The funding organizations played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
-
Data availability: Clinical data have been collected from patients in accordance with agreements and regulations for consent that prohibit transfer to third parties. Other datasets used or analyzed during the study are available from the corresponding author upon reasonable request.
References
1. Sung, H, Ferlay, J, Siegel, R, Laversanne, M, Soerjomataram, I, Jemal, A, et al.. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49.10.3322/caac.21660Search in Google Scholar PubMed
2. Mohandas, KM. Colorectal cancer in India: controversies, enigmas and primary prevention. Indian J Gastroenterol 2011;30:3–6. https://doi.org/10.1007/s12664-010-0076-2.Search in Google Scholar PubMed
3. Brenner, H, Kloor, M, Pox, CP. Colorectal cancer. Lancet 2014;383:1490–502. https://doi.org/10.1016/S0140-6736(13)61649-9.Search in Google Scholar PubMed
4. Bhurgri, Y, Khan, T, Kayani, N, Ahmad, R, Usman, A, Bhurgri, A, et al.. Incidence and current trends of colorectal malignancies in an unscreened, low risk Pakistan population. Asian Pac J Cancer Prev 2011;12:703–8.Search in Google Scholar
5. Efremidou, EI, Liratzopoulos, N, Papageorgiou, SM, Romanidis, K, Tourlis, T, Kouklakis, G, et al.. Colorectal carcinoma: correlation between age, gender and subsite distribution. Chirurgia 2008;103:659–63.Search in Google Scholar
6. Araghi, M, Soerjomataram, I, Bardot, A, Ferlay, J, Cabasag, CJ, Morrison, DS, et al.. Changes in colorectal cancer incidence in seven high-income countries: a population-based study. Lancet Gastroenterol Hepatol 2019;4:511–8. https://doi.org/10.1016/S2468-1253(19)30147-5.Search in Google Scholar PubMed
7. Amini, AQ, Samo, KA, Memon, AS. Colorectal cancer in younger population: our experience. J Pak Med Assoc 2013;63:1275–7.Search in Google Scholar
8. Kansakar, P, Singh, Y. Changing trends of colorectal carcinoma in Nepalese young adults. Asian Pac J Cancer Prev 2012;13:3209–12. https://doi.org/10.7314/apjcp.2012.13.7.3209.Search in Google Scholar PubMed
9. Feinberg, AP, Tycko, B. The history of cancer epigenetics. Nat Rev Cancer 2004;4:143–53. https://doi.org/10.1038/nrc1279.Search in Google Scholar PubMed
10. Kass, SU, Pruss, D, Wolffe, AP. How does DNA methylation repress transcription? Trends Genet 1997;13:444–9. https://doi.org/10.1016/s0168-9525(97)01268-7.Search in Google Scholar PubMed
11. Aaltonen, LA, Peltomaki, P, Leach, FS, Sistonen, P, Pylkkanen, L, Mecklin, JP, et al.. Clues to the pathogenesis of familial colorectal cancer. Science 1993;260:812–16. https://doi.org/10.1126/science.8484121.Search in Google Scholar PubMed
12. Herman, JG, Umar, A, Polyak, K, Graff, JR, Ahuja, N, Issa, JP, et al.. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A 1998;95:6870–5. https://doi.org/10.1073/pnas.95.12.6870.Search in Google Scholar PubMed PubMed Central
13. Ramirez, N, Bandres, E, Navarro, A, Pons, A, Jansa, S, Moreno, I, et al.. Epigenetic events in normal colonic mucosa surrounding colorectal cancer lesions. Eur J Cancer 2008;44:2689–95. https://doi.org/10.1016/j.ejca.2008.09.004.Search in Google Scholar PubMed
14. Ozenne, P, Eymin, B, Brambilla, E, Gazzeri, S. The ARF tumor suppressor: structure, functions and status in cancer. Int J Cancer 2010;127:2239–47. https://doi.org/10.1002/ijc.25511.Search in Google Scholar PubMed
15. Wettergren, Y, Odin, E, Nilsson, S, Carlsson, G, Gustavsson, B. p16INK4a gene promoter hypermethylation in mucosa as a prognostic factor for patients with colorectal cancer. Mol Med 2008;14:412–21. https://doi.org/10.2119/2007-00096.Wettergren.Search in Google Scholar PubMed PubMed Central
16. Yang, L, Xiong, Z, Xie, Q, He, W, Liu, S, Kong, P, et al.. Prognostic value of total number of lymph nodes retrieved differs between left-sided colon cancer and right-sided colon cancer in stage III patients with colon cancer. BMC Cancer 2018;18:558. https://doi.org/10.1186/s12885-018-4431-5.Search in Google Scholar PubMed PubMed Central
17. Elliott, GO, Johnson, IT, Scarll, J, Dainty, J, Williams, EA, Garg, D, et al.. Quantitative profiling of CpG island methylation in human stool for colorectal cancer detection. Int J Colorectal Dis 2013;28:35–42. https://doi.org/10.1007/s00384-012-1532-5.Search in Google Scholar PubMed
18. Abdul, QA, Yu, BP, Chung, HY, Jung, HA, Choi, JS. Epigenetic modifications of gene expression by lifestyle and environment. Arch Pharm Res 2017;40:1219–37. https://doi.org/10.1007/s12272-017-0973-3.Search in Google Scholar PubMed
19. Coppede, F, Migheli, F, Lopomo, A, Failli, A, Legitimo, A, Consolini, R, et al.. Gene promoter methylation in colorectal cancer and healthy adjacent mucosa specimens: correlation with physiological and pathological characteristics, and with biomarkers of one-carbon metabolism. Epigenetics 2014;9:621–33. https://doi.org/10.4161/epi.27956.Search in Google Scholar PubMed PubMed Central
20. Arai, T, Kasahara, I, Sawabe, M, Honma, N, Aida, J, Tabubo, K. Role of methylation of the hMLH1 gene promoter in the development of gastric and colorectal carcinoma in the elderly. Geriatr Gerontol Int 2010;10:S207-12. https://doi.org/10.1111/j.1447-0594.2010.00590.x.Search in Google Scholar PubMed
21. Menigatti, M, Truninger, K, Gebbers, JO, Marbet, U, Marra, G, Schar, P, et al.. Normal colorectal mucosa exhibits sex- and segment-specific susceptibility to DNA methylation at the hMLH1 and MGMT promoters. Oncogene 2009;28:899–909. https://doi.org/10.1038/onc.2008.444.Search in Google Scholar PubMed
22. Wiencke, JK, Zheng, S, Lafuente, A, Lafuente, MJ, Grudzen, C, Wrensch, MR, et al.. Aberrant methylation of p16INK4a in anatomic and gender-specific subtypes of sporadic colorectal cancer. Cancer Epidemiol Biomarkers Prev 1999;8:501–6.Search in Google Scholar
23. Kaz, AM, Wong, CJ, Dzieciatkowski, S, Luo, Y, Schoen, RE, Grady, WM. Patterns of DNA methylation in the normal colon vary by anatomical location, gender, and age. Epigenetics 2014;9:492–502. https://doi.org/10.4161/epi.27650.Search in Google Scholar PubMed PubMed Central
24. Slaughter, DP, Southwick, HW, Smejkal, W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 1953;6:963–8. https://doi.org/10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q.10.1002/1097-0142(195309)6:5<963::AID-CNCR2820060515>3.0.CO;2-QSearch in Google Scholar
25. Shen, L, Kondo, Y, Rosner, GL, Xiao, L, Hernandez, NS, Vilaythong, J, et al.. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst 2005;97:1330–8. https://doi.org/10.1093/jnci/dji275.Search in Google Scholar PubMed
26. Wettergren, Y, Odin, E, Carlsson, G, Gustavsson, B. MTHFR, MTR, and MTRR polymorphisms in relation to p16INK4A hypermethylation in mucosa of patients with colorectal cancer. Mol Med 2010;16:425–32. https://doi.org/10.2119/molmed.2009.00156.Search in Google Scholar PubMed PubMed Central
27. Ma, Z, Williams, M, Cheng, YY, Leung, WK. Roles of methylated DNA biomarkers in patients with colorectal cancer. Dis Markers 2019;2019:2673543. https://doi.org/10.1155/2019/2673543.Search in Google Scholar PubMed PubMed Central
28. Giddings, BH, Kwong, SL, Parikh-Patel, A, Bates, JH, Snipes, KP. Going against the tide: increasing incidence of colorectal cancer among Koreans, Filipinos, and South Asians in California, 1988-2007. Cancer Causes Control 2012;23:691–702. https://doi.org/10.1007/s10552-012-9937-6.Search in Google Scholar PubMed
29. Pan, P, Yu, J, Wang, LS. Diet and colon: what matters? Curr Opin Gastroenterol 2019;35:101–6. https://doi.org/10.1097/MOG.0000000000000501.Search in Google Scholar PubMed PubMed Central
30. Ayers, D, Boughanem, H, Macias-Gonzalez, M. Epigenetic influences in the obesity/colorectal cancer axis: a novel theragnostic avenue. J Oncol 2019;2019:7406078. https://doi.org/10.1155/2019/7406078.Search in Google Scholar PubMed PubMed Central
31. Ye, C, Shrubsole, MJ, Cai, Q, Ness, R, Grady, WM, Smalley, W, et al.. Promoter methylation status of the MGMT, hMLH1, and CDKN2A/p16 genes in non-neoplastic mucosa of patients with and without colorectal adenomas. Oncol Rep 2006;16:429–35.10.3892/or.16.2.429Search in Google Scholar
32. Neovius, M, Janson, A, Rossner, S. Prevalence of obesity in Sweden. Obes Rev 2006;7:1–3. https://doi.org/10.1111/j.1467-789x.2006.00190.x.Search in Google Scholar PubMed
33. Gutierrez-Angulo, M, Ayala-Madrigal, ML, Moreno-Ortiz, JM, Peregrina-Sandoval, J, Garcia-Ayala, FD. Microbiota composition and its impact on DNA methylation in colorectal cancer. Front Genet 2023;14:1037406. https://doi.org/10.3389/fgene.2023.1037406.Search in Google Scholar PubMed PubMed Central
34. Sun, A, Park, P, Cole, L, Vaidya, H, Maegawa, S, Keith, K, et al.. Non-pathogenic microbiota accelerate age-related CpG Island methylation in colonic mucosa. Epigenetics 2023;18:2160568. https://doi.org/10.1080/15592294.2022.2160568.Search in Google Scholar PubMed PubMed Central
35. Strum, WB, Boland, CR. Clinical and genetic characteristics of colorectal cancer in persons under 50 years of age: a review. Dig Dis Sci 2019;64:3059–65. https://doi.org/10.1007/s10620-019-05644-0.Search in Google Scholar PubMed
36. Hirai, HW, Tsoi, KK, Chan, JY, Wong, SH, Ching, JY, Wong, MC, et al.. Systematic review with meta-analysis: faecal occult blood tests show lower colorectal cancer detection rates in the proximal colon in colonoscopy-verified diagnostic studies. Aliment Pharmacol Ther 2016;43:755–64. https://doi.org/10.1111/apt.13556.Search in Google Scholar PubMed
37. Song, L, Jia, J, Peng, X, Xiao, W, Li, Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: a meta-analysis. Sci Rep 2017;7:3032. https://doi.org/10.1038/s41598-017-03321-8.Search in Google Scholar PubMed PubMed Central
38. Patil, PS, Saklani, A, Gambhire, P, Mehta, S, Engineer, R, De’Souza, A, et al.. Colorectal cancer in India: an audit from a tertiary center in a low prevalence area. Indian J Surg Oncol 2017;8:484–90. https://doi.org/10.1007/s13193-017-0655-0.Search in Google Scholar PubMed PubMed Central
39. Oliveira, D, Hentze, J, O’Rourke, CJ, Andersen, JB, Hogdall, C, Hogdall, EV. DNA Methylation in Ovarian Tumors-a Comparison Between Fresh Tissue and FFPE Samples. Reprod Sci 2021;28:3212–8.10.1007/s43032-021-00589-0Search in Google Scholar PubMed PubMed Central
40. Moran, B, Das, S, Smeets, D, Peutman, G, Klinger, R, Fender, B, et al.. Assessment of concordance between fresh-frozen and formalin-fixed paraffin embedded tumor DNA methylation using a targeted sequencing approach. Oncotarget 2017;8:48126–37.10.18632/oncotarget.18296Search in Google Scholar PubMed PubMed Central
Supplementary Material
The online version of this article offers supplementary material (https://doi.org/10.1515/iss-2023-0039).
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- Review
- Near-infrared indocyanine green angiography in recognizing bowel ischemia in emergency surgery: game changer or overrated?
- Original Articles
- Learning curves for high tibial osteotomy using patient-specific instrumentation: a case control study
- Biological conduits based on spider silk for reconstruction of extended nerve defects
- The effect of adipose-derived stem cells (ADSC) treatment on kidney histopathological appearance on the Wistar rat models with grade five kidney trauma
- Epigenetic differences in the tumor suppressor genes MLH1 and p16INK4a between Nepalese and Swedish patients with colorectal cancer
Articles in the same Issue
- Frontmatter
- Review
- Near-infrared indocyanine green angiography in recognizing bowel ischemia in emergency surgery: game changer or overrated?
- Original Articles
- Learning curves for high tibial osteotomy using patient-specific instrumentation: a case control study
- Biological conduits based on spider silk for reconstruction of extended nerve defects
- The effect of adipose-derived stem cells (ADSC) treatment on kidney histopathological appearance on the Wistar rat models with grade five kidney trauma
- Epigenetic differences in the tumor suppressor genes MLH1 and p16INK4a between Nepalese and Swedish patients with colorectal cancer

