Abstract
Background:
Clinical Text Analysis and Knowledge Extraction System (cTAKES) is an open-source natural language processing (NLP) system. In recent development modules of cTAKES, a negation detection (ND) algorithm is used to improve annotation capabilities and simplify automatic identification of negative context in large clinical documents. In this research, the two types of ND algorithms used are lexicon and syntax, which are analyzed using a database made openly available by the National Center for Biomedical Computing. The aim of this analysis is to find the pros and cons of these algorithms.
Methods:
Patient medical reports were collected from three institutions included the 2010 i2b2/VA Clinical NLP Challenge, which is the input data for this analysis. This database includes patient discharge summaries and progress notes. The patient data is fed into five ND algorithms: NegEx, ConText, pyConTextNLP, DEEPEN and Negation Resolution (NR). NegEx, ConText and pyConTextNLP are lexicon-based, whereas DEEPEN and NR are syntax-based. The results from these five ND algorithms are post-processed and compared with the annotated data. Finally, the performance of these ND algorithms is evaluated by computing standard measures including F-measure, kappa statistics and ROC, among others, as well as the execution time of each algorithm.
Results:
This research is tested through practical implementation based on the accuracy of each algorithm’s results and computational time to evaluate its performance in order to find a robust and reliable ND algorithm.
Conclusions:
The performance of the chosen ND algorithms is analyzed based on the results produced by this research approach. The time and accuracy of each algorithm are calculated and compared to suggest the best method.
Acknowledgments
We thank S. Shahul Hameed and Gokulalakshmi Elayaperumal from Sree Balaji Medical College and Hospital, Chennai, India, George Gkotsis from IoPPN, King’s College London, and Saeed Mehrabi from the School of Informatics and Computing, Indiana University, Indianapolis, IN, USA, who offered their continuous support in the implementation of this task.
Author contributions: The authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding: None declared.
Employment or leadership: None declared.
Honorarium: None declared.
Competing interests: The funding organization(s) played no role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
Appendix A
Example of single annotated i2b2 report.
| Sentences (report file) | Chest CT scan was negative for pulmonary embolism but positive for consolidation |
|---|---|
| Assertion annotation | c=“pulmonary embolism” 21:6 21:7 | | t= “problem” | | a=“absent” |
| (Annotated file) | c=“consolidation” 21:11 21:11 | | t=“problem” | | a=“present” |
A sample input data.
| Concepts | Sentences |
|---|---|
| Mass | No mass or vegetation is seen on the mitral valve |
| Pericardial effusion | There is no pericardial effusion |
| Epileptiform features | No epileptiform features were seen |
| Infection | CXR, LP, UA and abdominal CT showed no sign of infection |
| Orthostatic | She was not orthostatic |
| A headache | He did not complain about a headache |
Evaluation of the algorithm’s output using 2*2 table.
| Predicted output | ||
|---|---|---|
| True (negated) | False (affirmed) | |
| Manual annotations | ||
| True (negated) | True positive (TP) | False negative (FN) |
| False (affirmed) | False positive (FP) | True negative (TN) |
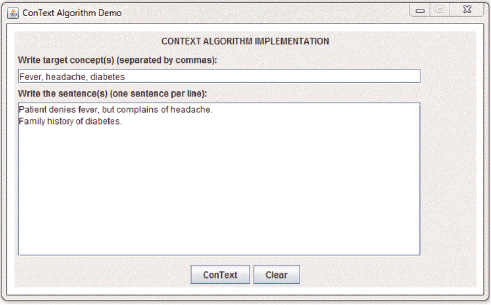
A simple surface-based approach of ConText algorithm.
References
1. Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc 2012;18:544–51.10.1136/amiajnl-2011-000464Suche in Google Scholar PubMed PubMed Central
2. Koopman B, Bruza P, Sitbon L, Lawley M. Analysis of the effect of negation on information retrieval of medical data. In: Proc 15th Australas Doc Comput Symp 2010:89–92.Suche in Google Scholar
3. Scuba W, Tharp M, Mowery D, Tseytlin E, Liu Y, Drews FA, et al. Knowledge author: facilitating user-driven, domain content development to support clinical information extraction. J Biomed Semant 2016;7:42.10.1186/s13326-016-0086-9Suche in Google Scholar PubMed PubMed Central
4. Garla V, Re V Lo, Dorey-Stein Z, Kidwai F, Scotch M, Womack J, et al. The Yale cTAKES extensions for document classification: architecture and application. J Am Med Inform Assoc 2011;18:614–20.10.1136/amiajnl-2011-000093Suche in Google Scholar PubMed PubMed Central
5. Mitchell KJ, Becich MJ, Berman JJ, Chapman WW, Gilbertson J, Gupta D, et al. Implementation and evaluation of a negation tagger in a pipeline-based system for information extraction from pathology reports. Stud Health Technol Inform 2004;107:663–7.Suche in Google Scholar
6. Clark C, Aberdeen J, Coarr M, Tresner-kirsch D, Wellner B, Yeh A, et al. Determining assertion status for medical problems in clinical records. McLean, VA: Mitre Corporation, 2011:2–6.Suche in Google Scholar
7. Ou Y, Patrick J. Automatic negation detection in narrative pathology reports. Artif Intell Med 2015;64:41–50.10.1016/j.artmed.2015.03.001Suche in Google Scholar PubMed
8. Jiang M, Chen Y, Liu M, Rosenbloom ST, Mani S, Denny JC, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inf Assoc 2011;18:601–6.10.1136/amiajnl-2011-000163Suche in Google Scholar PubMed PubMed Central
9. Clark C, Aberdeen J, Coarr M, Tresner-Kirsch D, Wellner B, Yeh A, et al. MITRE system for clinical assertion status classification. J Am Med Inform Assoc 2011;18:563–7.10.1136/amiajnl-2011-000164Suche in Google Scholar PubMed PubMed Central
10. Minard A-L, Ligozat A-L, Ben Abacha A, Bernhard D, Cartoni B, Deléger L, et al. Hybrid methods for improving information access in clinical documents: concept, assertion, and relation identification. J Am Med Inform Assoc 2011;18:588–93.10.1136/amiajnl-2011-000154Suche in Google Scholar PubMed PubMed Central
11. Ballesteros M, Francisco V, Díaz A, Herrera J, Gervás P. Inferring the scope of negation in biomedical documents. Lect Notes Comput Sci 2012;7181 LNCS:363–75.10.1007/978-3-642-28604-9_30Suche in Google Scholar
12. Chapman WW, Dowling JN, Wagner MM. Fever detection from free-text clinical records for biosurveillance. J Biomed Inform 2004;37:120–7.10.1016/j.jbi.2004.03.002Suche in Google Scholar PubMed PubMed Central
13. Sanchez-Graillet O, Poesio M. Negation of protein-protein interactions: analysis and extraction. Bioinformatics 2007;23:424–32.10.1093/bioinformatics/btm184Suche in Google Scholar PubMed
14. Morante R. Descriptive analysis of negation cues in biomedical texts. Statistics 2009;1429–36.Suche in Google Scholar
15. Horn LR. Natural history of negation. J Pragmat 1989;16: 269–80.10.1016/0378-2166(91)90096-GSuche in Google Scholar
16. Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10.10.1006/jbin.2001.1029Suche in Google Scholar PubMed
17. Aronow DB, Fangfang F, Croft WB. Ad hoc classification of radiology reports. J Am Med Inform Assoc 1999;6:393–411.10.1136/jamia.1999.0060393Suche in Google Scholar PubMed PubMed Central
18. Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc 2001;8:598–609.10.1136/jamia.2001.0080598Suche in Google Scholar PubMed PubMed Central
19. Gindl S, Kaiser K, Miksch S. Syntactical negation detection in clinical practice guidelines. Stud Health Technol Inform 2008;136:187–92.Suche in Google Scholar PubMed
20. Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009;42:839–51.10.1016/j.jbi.2009.05.002Suche in Google Scholar PubMed PubMed Central
21. Chapman BE, Lee S, Kang HP, Chapman WW. Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm. J Biomed Inform 2011;44:728–37.10.1016/j.jbi.2011.03.011Suche in Google Scholar PubMed PubMed Central
22. Mehrabi S, Krishnan A, Sohn S, Roch AM, Schmidt H, Kesterson J, et al. DEEPEN: a negation detection system for clinical text incorporating dependency relation into NegEx. J Biomed Inform 2015;54:213–9.10.1016/j.jbi.2015.02.010Suche in Google Scholar PubMed PubMed Central
23. Huang Y, Lowe H. A novel hybrid approach to automated negation detection in clinical radiology reports. J Am Med Inform 2007;304–11.10.1197/jamia.M2284Suche in Google Scholar PubMed PubMed Central
24. Zhu Q, Li J, Wang H. A unified framework for scope learning via simplified shallow semantic parsing. In: EMNLP 2010 Proceedings of the 2010 Conference on Empirical Methods in Natural Language Processing 2010:714–24.Suche in Google Scholar
25. Sohn S, Wu S, Chute CG. Dependency parser-based negation detection in clinical narratives. AMIA Jt Summits Transl Sci Proc AMIA Summit Transl Sci 2012;2012:1–8.Suche in Google Scholar
26. Gkotsis G, Velupillai S, Oellrich A, Dean H, Liakata M, Dutta R. Don’t let notes be misunderstood: a negation detection method for assessing risk of suicide in mental health records. In: Proc 3rd Work Comput Linguist Clin Psychol Linguist Signal Clin Real 2016:95–105.10.18653/v1/W16-0310Suche in Google Scholar
27. Lapponi E, Read J, Øvrelid L. Representing and resolving negation for sentiment analysis. In: Proc 12th IEEE Int Conf Data Min Work ICDMW 2012:687–92.10.1109/ICDMW.2012.23Suche in Google Scholar
28. Shivade C, de Marneffe MC, Fosler-Lussier E, Lai AM. Extending NegEx with kernel methods for negation detection in clinical text. In: Proc Work Extra-Propositional Asp Mean Comput Semant NAACL 2015:41–6.10.3115/v1/W15-1305Suche in Google Scholar
29. Kang T, Zhang S, Xu N, Wen D, Zhang X, Lei J. Detecting negation and scope in Chinese clinical notes using character and word embedding. Comput Methods Programs Biomed 2017;140:53–9.10.1016/j.cmpb.2016.11.009Suche in Google Scholar PubMed
30. Goryachev S, Sordo M, Zeng QT, Ngo L. Implementation and evaluation of four different methods of negation detection. Boston, MA: DSG, 2006.Suche in Google Scholar
31. Tanushi H, Dalianis H, Duneld M, Kvist M, Skeppstedt M, Velupillai S. Negation scope delimitation in clinical text using three approaches: NegEx, PyConTextNLP and SynNeg. In: Proc 19th Nord Conf Comput Linguist (NoDaLiDa 2013) 2013;1: 387–97.Suche in Google Scholar
32. Wu S, Miller T, Masanz J, Coarr M, Halgrim S, Carrell D, et al. Negation’s not solved: generalizability versus optimizability in clinical natural language processing. PLoS One 2014;9:e112774.10.1371/journal.pone.0112774Suche in Google Scholar PubMed PubMed Central
33. Uzuner O, South BR, Shen S, DuVall SL, Uzuner Ö, South BR, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6.10.1136/amiajnl-2011-000203Suche in Google Scholar PubMed PubMed Central
34. Mowery DL, Chapman BE, Conway M, South BR, Madden E, Keyhani S, et al. Extracting a stroke phenotype risk factor from Veteran Health Administration clinical reports: an information content analysis. J Biomed Semantics 2016;7:26.10.1186/s13326-016-0065-1Suche in Google Scholar PubMed PubMed Central
35. Chapman BE, Mowery DL, Narasimhan E, Patel N, Chapman WW, Heilbrun ME. Assessing the feasibility of an automated suggestion system for communicating critical findings from chest radiology reports to referring physicians. In: Proc 15th Work Biomed Nat Lang Process 2016:181–5.10.18653/v1/W16-2924Suche in Google Scholar
36. Bruha I, Famili A. Postprocessing in machine learning and data mining. ACM SIGKDD Explor Newslett 2000;2:110–4.10.1145/380995.381059Suche in Google Scholar
©2017 Walter de Gruyter GmbH, Berlin/Boston
Artikel in diesem Heft
- Frontmatter
- Research Articles
- Mechanism of ligand binding – PDZ domain taken as example
- Chain-chain complexation and heme binding in haemoglobin with respect to the hydrophobic core structure
- Modified S-transform as a tool to identify secondary structure elements in RNA
- Electronic health record for elderly patients
- Evaluation of lexicon- and syntax-based negation detection algorithms using clinical text data
- Short Communication
- An improved structural model of the human iron exporter ferroportin. Insight into the role of pathogenic mutations in hereditary hemochromatosis type 4
Artikel in diesem Heft
- Frontmatter
- Research Articles
- Mechanism of ligand binding – PDZ domain taken as example
- Chain-chain complexation and heme binding in haemoglobin with respect to the hydrophobic core structure
- Modified S-transform as a tool to identify secondary structure elements in RNA
- Electronic health record for elderly patients
- Evaluation of lexicon- and syntax-based negation detection algorithms using clinical text data
- Short Communication
- An improved structural model of the human iron exporter ferroportin. Insight into the role of pathogenic mutations in hereditary hemochromatosis type 4


