The crystal structure of poly[di(μ2-aqua)-diaqua-bis(3-aminopyridine-4-carboxylate-κ2 O: O′)-tetra(μ2-3-aminopyridine-4-carboxylate-κ2 O: O′)-dineodymium(III), [Nd2(C6H5N2O2)6(H2O)4] n
Abstract
[Nd2(C6H5N2O2)6(H2O)4] n , monoclinic, P21/c (no. 14), a = 9.383(3) Å, b = 22.741(6) Å, c = 18.775(5) Å, β = 90.231(5)°, V = 4006(2) Å3, Z = 4, Rgt (F) = 0.0427, wRref (F 2) = 0.0862, T = 296 K.
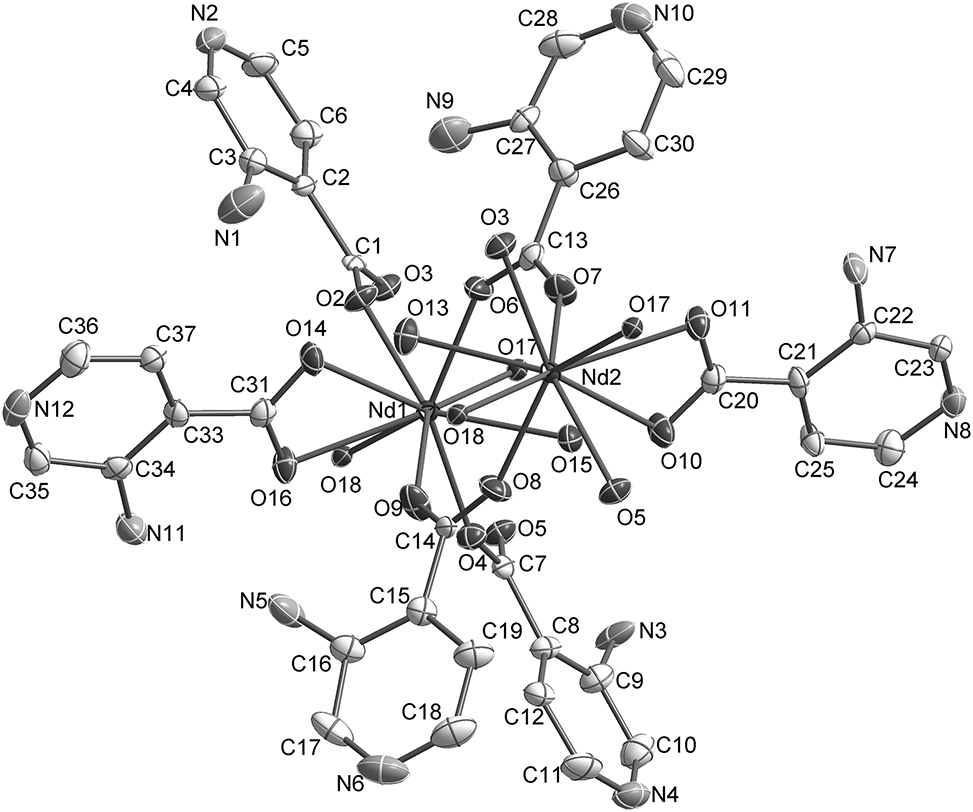
The molecular structure is shown in the figure. Table 1 contains crystallographic data and Table 2 contains the list of the atoms including atomic coordinates and displacement parameters.
Data collection and handling.
| Crystal: | Colorless block |
| Size: | 0.20 × 0.18 × 0.10 mm |
| Wavelength: | Mo Kα radiation (0.71073 Å) |
| μ: | 2.66 mm−1 |
| Diffractometer, scan mode: | Bruker APEX-II, φ and ω |
| θ max, completeness: | 27.6°, >99 % |
| N(hkl)measured, N(hkl)unique, R int: | 24,277, 9122, 0.063 |
| Criterion for I obs, N(hkl)gt: | I obs > 2 σ(I obs), 6092 |
| N(param)refined: | 605 |
| Programs: | Bruker [1], Diamond [2], Olex2 [3], SHELX [4, 5] |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2).
| Atom | x | y | z | U iso*/U eq |
|---|---|---|---|---|
| Nd1 | 0.49211 (3) | 0.35460 (2) | 0.25256 (2) | 0.01435 (8) |
| Nd2 | 1.04449 (3) | 0.39830 (2) | 0.26271 (2) | 0.01375 (8) |
| O17 | 0.3219 (3) | 0.43644 (14) | 0.28824 (19) | 0.0190 (8) |
| H17A | 0.332005 | 0.444117 | 0.332611 | 0.028* |
| H17B | 0.336265 | 0.465857 | 0.263161 | 0.028* |
| O2 | 0.3641 (4) | 0.32326 (16) | 0.3573 (2) | 0.0227 (9) |
| O3 | 0.1471 (4) | 0.36036 (16) | 0.3721 (2) | 0.0223 (9) |
| O4 | 0.3895 (4) | 0.38804 (16) | 0.14489 (19) | 0.0219 (9) |
| O5 | 0.1729 (4) | 0.42775 (16) | 0.1527 (2) | 0.0240 (9) |
| O6 | 0.6358 (4) | 0.38627 (16) | 0.3514 (2) | 0.0248 (9) |
| O7 | 0.8481 (4) | 0.42707 (17) | 0.3392 (2) | 0.0288 (10) |
| O8 | 0.8946 (4) | 0.36458 (17) | 0.1701 (2) | 0.0280 (10) |
| O9 | 0.6796 (4) | 0.32430 (17) | 0.1688 (2) | 0.0305 (10) |
| O10 | 0.9188 (4) | 0.48624 (16) | 0.2102 (2) | 0.0278 (10) |
| O11 | 1.0712 (4) | 0.50565 (15) | 0.2961 (2) | 0.0239(10) |
| O18 | 1.2009 (3) | 0.31453 (14) | 0.22682 (19) | 0.0196 (8) |
| H18A | 1.190447 | 0.306198 | 0.183953 | 0.029* |
| H18B | 1.183987 | 0.282718 | 0.252823 | 0.029* |
| O13 | 0.9118 (4) | 0.30711 (15) | 0.3059 (2) | 0.0242 (9) |
| H13A | 0.966630 | 0.278136 | 0.303797 | 0.036* |
| H13B | 0.827120 | 0.305276 | 0.287157 | 0.036* |
| O14 | 0.6150 (4) | 0.26242 (15) | 0.3004 (2) | 0.0250 (9) |
| O15 | 0.6233 (3) | 0.44527 (15) | 0.2172 (2) | 0.0222 (9) |
| H15A | 0.663738 | 0.471007 | 0.240164 | 0.033* |
| H15B | 0.611437 | 0.451907 | 0.171814 | 0.033* |
| N1 | 0.4474 (5) | 0.2346 (2) | 0.4431 (3) | 0.0498 (17) |
| H1A | 0.500895 | 0.206943 | 0.459449 | 0.060* |
| H1B | 0.463952 | 0.249337 | 0.401782 | 0.060* |
| N2 | 0.2119 (5) | 0.2420 (2) | 0.5932 (3) | 0.0292 (12) |
| N3a | 0.1029 (9) | 0.5117 (4) | 0.0600 (4) | 0.036 (3) |
| H3Aa | 0.055540 | 0.540824 | 0.042599 | 0.044* |
| H3Ba | 0.082424 | 0.498311 | 0.101494 | 0.044* |
| N4 | 0.3488 (5) | 0.4930 (2) | −0.0866 (3) | 0.0318 (13) |
| N5 | 0.6720 (5) | 0.2251 (2) | 0.0826 (3) | 0.0416 (15) |
| H5A | 0.653680 | 0.234067 | 0.126133 | 0.050* |
| H5B | 0.633608 | 0.194446 | 0.063716 | 0.050* |
| N6 | 0.8905 (6) | 0.2677 (3) | −0.0666 (3) | 0.0455 (16) |
| N7 | 1.1154 (5) | 0.61980 (19) | 0.3327 (3) | 0.0278 (12) |
| H7A | 1.135478 | 0.584279 | 0.344883 | 0.033* |
| H7B | 1.156977 | 0.648773 | 0.353677 | 0.033* |
| N8 | 0.8775 (5) | 0.70374 (19) | 0.2196 (3) | 0.0258 (12) |
| N9 | 0.5324 (5) | 0.3836 (3) | 0.4886 (3) | 0.0489 (16) |
| H9A | 0.474985 | 0.374649 | 0.522530 | 0.059* |
| H9B | 0.533580 | 0.362783 | 0.450327 | 0.059* |
| N10 | 0.6961 (6) | 0.5074 (2) | 0.5742 (3) | 0.0402 (15) |
| N11 | 0.3938 (5) | 0.1409 (2) | 0.1704 (3) | 0.0329 (13) |
| H11A | 0.349967 | 0.114278 | 0.146312 | 0.039* |
| H11B | 0.375878 | 0.177462 | 0.162787 | 0.039* |
| N12 | 0.6206 (5) | 0.0448 (2) | 0.2743 (3) | 0.0293 (12) |
| C1 | 0.2514 (5) | 0.3300 (2) | 0.3918 (3) | 0.0157 (12) |
| C2 | 0.2404 (6) | 0.2990 (2) | 0.4618 (3) | 0.0185 (12) |
| C3 | 0.3371 (6) | 0.2548 (2) | 0.4824 (3) | 0.0209 (13) |
| C4 | 0.3159 (6) | 0.2291 (3) | 0.5487 (3) | 0.0269 (14) |
| H4 | 0.380522 | 0.200365 | 0.562882 | 0.032* |
| C5 | 0.1210 (6) | 0.2839 (3) | 0.5719 (3) | 0.0296 (15) |
| H5 | 0.046426 | 0.293797 | 0.602089 | 0.036* |
| C6 | 0.1314 (6) | 0.3127 (3) | 0.5087 (3) | 0.0253 (14) |
| H6 | 0.065431 | 0.341665 | 0.496927 | 0.030* |
| C7 | 0.2865 (6) | 0.4183 (2) | 0.1197 (3) | 0.0179 (12) |
| C8 | 0.3049 (6) | 0.4428 (2) | 0.0468 (3) | 0.0204 (13) |
| C9 | 0.2134 (6) | 0.4862 (3) | 0.0205 (3) | 0.0272 (14) |
| H9b | 0.135727 | 0.499106 | 0.046892 | 0.033* |
| C10 | 0.2420 (7) | 0.5094 (3) | −0.0459 (4) | 0.0351 (16) |
| H10 | 0.181564 | 0.538634 | −0.063087 | 0.042* |
| C11 | 0.4343 (6) | 0.4511 (3) | −0.0616 (3) | 0.0348 (16) |
| H11 | 0.509144 | 0.438386 | −0.090008 | 0.042* |
| C12 | 0.4173 (6) | 0.4258 (3) | 0.0041 (3) | 0.0243 (14) |
| H12a | 0.481224 | 0.397333 | 0.019876 | 0.029* |
| C13 | 0.7369 (6) | 0.4174 (2) | 0.3733 (3) | 0.0190 (12) |
| C14 | 0.7974 (6) | 0.3347 (2) | 0.1421 (3) | 0.0174 (12) |
| C15 | 0.8287 (5) | 0.3098 (2) | 0.0700 (3) | 0.0200 (12) |
| C16 | 0.7620 (6) | 0.2595 (3) | 0.0435 (3) | 0.0243 (13) |
| C17 | 0.7982 (7) | 0.2409 (3) | −0.0258 (4) | 0.0373 (17) |
| H17 | 0.753572 | 0.207506 | −0.043628 | 0.045* |
| C18 | 0.9557 (7) | 0.3152 (3) | −0.0399 (4) | 0.0413 (17) |
| H18 | 1.022357 | 0.334542 | −0.067942 | 0.050* |
| C19 | 0.9289 (6) | 0.3366 (3) | 0.0267 (3) | 0.0323 (15) |
| H19 | 0.978297 | 0.369447 | 0.043111 | 0.039* |
| C20 | 0.9793 (6) | 0.5221 (2) | 0.2500 (3) | 0.0197 (13) |
| C21 | 0.9451 (5) | 0.5865 (2) | 0.2433 (3) | 0.0192 (13) |
| C22 | 1.0182 (6) | 0.6301 (2) | 0.2805 (3) | 0.0206 (13) |
| C23 | 0.9805 (6) | 0.6886 (2) | 0.2635 (3) | 0.0234 (13) |
| H23 | 1.032371 | 0.718655 | 0.284970 | 0.028* |
| C24 | 0.8056 (6) | 0.6609 (3) | 0.1858 (3) | 0.0281 (14) |
| H24 | 0.731770 | 0.670806 | 0.154862 | 0.034* |
| C25 | 0.8390 (5) | 0.6025 (2) | 0.1960 (3) | 0.0221 (13) |
| H25 | 0.789617 | 0.573695 | 0.170927 | 0.027* |
| C26 | 0.7216 (5) | 0.4468 (2) | 0.4443 (3) | 0.0221 (13) |
| C27 | 0.6210 (5) | 0.4304 (2) | 0.4954 (3) | 0.0197 (13) |
| C28 | 0.6141 (7) | 0.4621 (3) | 0.5581 (3) | 0.0361 (16) |
| H28 | 0.546609 | 0.450637 | 0.591448 | 0.043* |
| C29 | 0.7917 (8) | 0.5230 (3) | 0.5259 (4) | 0.0449 (19) |
| H29 | 0.851187 | 0.554587 | 0.536135 | 0.054* |
| C30 | 0.8077 (6) | 0.4950 (3) | 0.4614 (3) | 0.0343 (16) |
| H30 | 0.875733 | 0.508203 | 0.429238 | 0.041* |
| C31 | 0.5475 (6) | 0.2296 (2) | 0.2586 (3) | 0.0200 (13) |
| O16 | 0.4587 (4) | 0.25024 (15) | 0.2148 (2) | 0.0241 (10) |
| C33 | 0.5704 (5) | 0.1648 (2) | 0.2622 (3) | 0.0190 (12) |
| C34 | 0.4915 (6) | 0.1252 (2) | 0.2208 (3) | 0.0199 (13) |
| C35 | 0.5183 (6) | 0.0655 (3) | 0.2320 (3) | 0.0278 (14) |
| H35 | 0.460816 | 0.038411 | 0.208341 | 0.033* |
| C36 | 0.6973 (6) | 0.0833 (3) | 0.3112 (4) | 0.0305 (15) |
| H36 | 0.769784 | 0.069407 | 0.340718 | 0.037* |
| C37 | 0.6739 (6) | 0.1422 (2) | 0.3075 (3) | 0.0221 (13) |
| H37 | 0.727830 | 0.167549 | 0.335649 | 0.027* |
| N3Ab | 0.5295 (12) | 0.3863 (5) | 0.0206 (6) | 0.030 (4) |
| H3AAb | 0.597268 | 0.380750 | −0.009397 | 0.036* |
| H3ABb | 0.529864 | 0.367833 | 0.060574 | 0.036* |
-
aOccupancy: 0.596(9), bOccupancy: 0.404(9).
1 Source of materials
3-Aminoisonicotinic acid (0.6 mmol 0.083 g), Nd2O3 (0.2 mmol 0.067 g), and 0.4 mol L−1 NaOH solution (1 mL), were dissolved in deionized water (7 mL). The mixture was sealed in a 25-mL Teflon-lined steel autoclave, and heated at 453 K for 8 days, then slowly cooled to room temperature. Colorless crystals of title complex could be found in the product.
2 Experimental details
All hydrogen atoms were placed in calculated positions and refined as riding atoms. The values were set to be 1.2U eq or 1.5U eq of the parent atoms.
3 Comment
Lanthanide complexes of 3-aminoisonicotinic acid have attracted some attentions during recent decades [6–11]. However, transition metal complexes for this ligand are relatively rare. A complex ([Nd2(C6H5N2O2)6(H2O)4] n ) of 3-aminoisonicotinic acid has been prepared under solvothermal reaction conditions. The asymmetric unit of title complex contains one Nd13+ ion, one Nd23+ ion, six L− (HL = 3-aminoisonicotinic acid) ligands and four coordinated aqua molecules. Each Nd1(III) ion is nine-coordinated by four oxygen atoms of four bridging L− ligands, two chelating oxygen atoms of one L− ligand and three oxygen atoms of three coordinated aqua molecules. The environment of the Nd23+ ion is similar to Nd13+. Each of Nd2(III) ions is surrounded by two chelating oxygen atoms of a L− ligand, four oxygen atoms of four μ 2-bridging L− ligands and three water oxygen atoms, forming a nine-coordination geometry, too. The Nd–O distances vary from 2.359(4) to 2.918(3) Å [12], [13], [14]. The O–Nd1–O angles are in the range 51.35(12)–145.45(13)°. Among the six L− ligands of the asymmetric unit, four act as μ 2-bridges, linking Nd1(III) and Nd2(III) ions, and two chelate to Nd(III) ions. While two of the four coordinated aqua molecules also play the roles of bridges, connecting Nd1(III) and Nd2(III) ions, and the other two act as monodentate ligands.
Each Nd1(III) ion is connected to one Nd2(III) ion via two μ 2–L− and two water molecules, and to another Nd2(III) ion by two μ 2–L−–L ligands, forming the 1–D zigzag-chain structure of the title complex with nonbonding Nd····Nd separations of 4.32–5.28 Å and Nd–Nd–Nd angles of 155.408(11)°. The zigzag chains stack parallel along the b axis, constructing the 3–D architecture of the title complex.
Funding source: NSF (Natural Science Foundation) of Anhui Province
Award Identifier / Grant number: 2108085MB53
Funding source: The NSF for Distinguished Young Scholars of Anhui University
Award Identifier / Grant number: 2022AH020087
Funding source: University NSF of Anhui Province, Research
Award Identifier / Grant number: KJ2020A0647
-
Conflict of interest statement: The authors declare no conflicts of interest regarding this article.
-
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
-
Research funding: NSF (Natural Science Foundation) of Anhui Province (2108085MB53), the NSF for Distinguished Young Scholars of Anhui University (2022AH020087), University NSF of Anhui Province (KJ2020A0647), Research.
References
1. Bruker. SAINT, APEX3 and SADABS; Bruker AXS Inc.: Madison, WI, USA, 2013.Search in Google Scholar
2. Brandenburg, K. DIAMOND. Visual Crystal Structure Information System. Ver. 4.6.8; Crystal Impact GbR: Bonn, Germany, 2022.Search in Google Scholar
3. Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K., Puschmann, H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341; https://doi.org/10.1107/s0021889808042726.Search in Google Scholar
4. Sheldrick, G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8; https://doi.org/10.1107/s2053229614024218.Search in Google Scholar PubMed PubMed Central
5. Sheldrick, G. M. SHELXTL - integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, A71, 3–8.10.1107/S2053273314026370Search in Google Scholar PubMed PubMed Central
6. Xue, X., Shao, Z., Cui, Y., Wen, T., Chen, J., Geng, K., Mi, L., Hou, H. Water-stable amino-functionalized coordination polymer for efficient Hg2+capture. Cryst. Growth Des. 2022, 22, 1412–1420; https://doi.org/10.1021/acs.cgd.1c01370.Search in Google Scholar
7. Ghosh, D., Fukushima, T., Kobayashi, K., Sen, S., Kitagawa, S., Katod, T., Tanaka, K. Base assisted C -C coupling between carbonyl and polypyridyl ligands in a Ru -NADH-type carbonyl complex. Dalton Trans. 2017, 46, 4373–4381; https://doi.org/10.1039/c7dt00312a.Search in Google Scholar PubMed
8. Ruan, M., Li, A., Wen, Y., Zhou, L., Zhang, J., Xuan, X. Adenine-based Bio -MOFs with high water and acid? Base stability for ammonia capture. CrystEngComm 2022, 24, 7420–7426; https://doi.org/10.1039/d2ce01092e.Search in Google Scholar
9. Gantzler, N., Kim, M.-B., Robinson, A., Terban, M. W., Ghose, S., Dinnebier, R. E., York, A. H., Tiana, D., Simon, C. M., Thallapally, P. K. Computation-informed optimization of Ni(PyC)2 functionalization for noble gas separations. Cell. Rep. Phys. Sci. 2022, 3, 101025; https://doi.org/10.1016/j.xcrp.2022.101025.Search in Google Scholar
10. Alduhaish, O., Li, B., Arman, H., Lin, R.-B., Zhao, J. C.-G., Chen, B. A Two-dimensional microporous metal -organic framework for highly selective adsorption of carbon dioxide and acetylene. Chinese Chem. Lett. 2017, 28, 1653–1658; https://doi.org/10.1016/j.cclet.2017.04.025.Search in Google Scholar
11. Liu, F.-F., Ping, Z.-Y., Zhou, W.-W., Zhao, W. The crystal structure of poly[(μu2-3-Aminopyridine-4-Carboxylate-κ2Nu: O)2Zinc(II)], [Zn(C6H5N2O2)2]n. Z. Kristallogr. N. Cryst. Struct. 2023, 238, 1131–1132; https://doi.org/10.1515/ncrs-2023-0358.Search in Google Scholar
12. Tiihonen, A., Lahtinen, M. In-depth structural analysis of lanthanoid coordination networks based on a flexible tripodal zwitterionic isonicotinate ligand. CrystEngComm 2019, 21, 2286–2302; https://doi.org/10.1039/c8ce01015c.Search in Google Scholar
13. Mondal, K. C., Sengupta, O., Dutta, P., Seehra, M., Nayak, S. K., Mukherjee, P. S. Three-dimensional 3d-4f heterometallic polymers containing both azide and carboxylate as co-LIGANDS. Inorg. Chim. Acta 2009, 362, 1913–1917; https://doi.org/10.1016/j.ica.2008.09.018.Search in Google Scholar
14. Feng, R., Chen, L., Chen, Q.-H., Shan, X.-C., Gai, Y.-L., Jiang, F.-L., Hong, M.-C. Novel luminescent three-dimensional heterometallic complexes with 2-fold interpenetrating (3,6)-connected nets. Cryst. Growth Des. 2011, 11, 1705–1712; https://doi.org/10.1021/cg101642j.Search in Google Scholar
© 2024 the author(s), published by De Gruyter, Berlin/Boston
This work is licensed under the Creative Commons Attribution 4.0 International License.
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of tris((Z)-2-hydroxy-N-((E)-pyridin-2-ylmethylene)benzohydrazonato-k2O,N)europium(III), C39H30N9O6Eu
- Crystal structure of (E)-3-(benzylideneamino)-2-phenylthiazolidin-4-one, C16H14N2OS
- The crystal structure of (E)-4-fluoro-N′-(1-(o-tolyl)ethylidene)benzohydrazide, C16H15FN2O
- Crystal structure of (6-chloropyridin-3-yl)methyl 2-(6-methoxynaphthalen-2-yl)propanoate, C20H18ClNO3
- Crystal structure of methyl 3-methoxy-4-(2-methoxy-2-oxoethoxy)benzoate, C12H14O6
- The crystal structure of bis[(4-methoxyphenyl)(picolinoyl)amido-κ2 N:N′]copper(II), C26H22CuN4O4
- The crystal structure of poly[di(μ2-aqua)-diaqua-bis(3-aminopyridine-4-carboxylate-κ2 O: O′)-tetra(μ2-3-aminopyridine-4-carboxylate-κ2 O: O′)-dineodymium(III), [Nd2(C6H5N2O2)6(H2O)4] n
- The crystal structure of t-butyl 7-[3-(4-fluorophenyl)-1-(propan-2-yl)-1H-indol-2-yl]-3,5-dihydroxyhept-6-enoate, C28H34FNO4
- Crystal structure of catena-poly[(benzylamine-κ1 N)-(sorbato-κ1 O)-(μ2-sorbato-κ2 O,O′)-copper(II), C19H23CuNO4
- Crystal structure of (4-(2-chlorophenyl)-1H-pyrrol-3-yl)(ferrocenyl) methanone, C21H16ClFeNO
- The crystal structure of N-[4-(4-bromophenyl)-1,3-thiazol-2-yl]-3-(2-methylphenyl)-2-sulfanylprop-2-enamide hydrate, C19H17BrN2O2S2
- The crystal structure of N′-{5-[2-(2,6-dimethylphenoxy) acetamido]-4-hydroxy-1,6-diphenylhexan-2-yl}-3-methyl-2-(2-oxo-1,3-diazinan-1-yl)butanamide hydrate
- Crystal structure of 2-(naphthalen-1-yl)ethyl 2-(6-methoxynaphthalen-2-yl)propanoate, C26H24O3
- Crystal structure of naphthalen-1-ylmethyl 2-(6-methoxynaphthalen-2-yl)propanoate, C25H22O3
- Crystal structure of poly[diaqua- (μ4-5-(1H-1,2,4-triazol-1-yl)benzene-1,3-dicarboxylato-κ5N:O,O’:O’’:O’’’)calcium(II), C10H9CaN3O6
- Crystal structure of (E)-N′-(4-((E)-3-(dimethylamino)acryloyl)-3-hydroxyphenyl)-N, N-dimethylformimidamide, C14H19N3O2
- Crystal structure of (E)-3-(dimethylamino)-1-(2-hydroxy-4,6-dimethoxyphenyl)prop-2-en-1-one, C13H17NO4
- Crystal structure of (2-chloropyridin-3-yl)methyl-2-(6-methoxynaphthalen-2-yl)propanoate, C20H18ClNO3
- The crystal structure of diethyl 4-(3,4-dimethylphenyl)-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate, C21H27NO4
- Crystal structure of (8R,9S,10R,13S,14S,17S)-17-hydroxy-10,13-dimethyl-17-((4-(2-phenylpropyl)phenyl)ethynyl)-1,2,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-3H-cyclopenta[a]phenanthren-3-one, C36H42O2
- Synthesis and crystal structure of 4-(4-cyclopropylnaphthalen-1-yl)-2,4-dihydro-3H-1,2,4-triazole-3-thione, C15H13N3S
- Crystal structure of catena-poly[aqua-(2,6-di-(2-pyridyl)-pyridine-κ3 N,N′, N″)(μ2-1,4-naphthalene dicarboxylato-κ2 O,O′)nickel(II)], C27H19NiN3O5
- Crystal structure of 3-(diphenylphosphoryl)-3-hydroxy-1-phenylpropan-1-one, C21H19O3P
- The crystal structure of R,S-{N-[(2-oxidonaphthalen-1-yl)methylidene]phenylglycinato}divinylsilicon, C23H19NO3Si
- The crystal structure of 1,2,4-tris(bromomethyl)benzene, C9H9Br3
- Crystal structure of chlorido-[4-(pyridin-2-yl)benzaldehyde-κ2 N,C]-(diethylamine-κ1 N)platinum(II), C16H18ClN2OPt
- Crystal structure of 3-(methoxycarbonyl)-1-(4-methoxyphenyl)-2,3,4,9- tetrahydro-1H-pyrido[3,4-b]indol-2-ium chloride hydrate, C40H48Cl2N4O9
- The crystal structure of 1-(2-chlorobenzyl)-3-(3-chlorophenyl)urea, C14H12Cl2N2O
- Hydrothermal synthesis and crystal structure of aqua-tris(4-acetamidobenzoato-κ2 O,O′)-(1,10-phenanthroline-κ2 N,N′)terbium(III) hydrate C39H36N5O11Tb
- The crystal structure of zwitterionic 3-aminoisonicotinic acid, C6H6N2O2
- The crystal structure of bis{[monoaqua-μ2-4-[(pyridine-4-carbonyl)-amino]-phthalato-κ3 N:O,O′-(2,2′-bipyridine κ2 N,N′)copper(II)]}decahydrate, C48H56N8O22Cu2
- Crystal structure of poly[μ10-4,4′-methylene-bis(oxy)benzoatodipotassium], C15H10K2O6
- The crystal structure of catena-poly[[tetraaqua[(μ2-1,4-di(4-methyl-1-imidazolyl)benzene] cobalt(II)]bis(formate)], C16H24CoN4O8
- The crystal structure of (E)-2-chloro-5-((2-(nitromethylene)imidazolidin-1-yl)methyl)pyridine, C10H11ClN4O2
- The crystal structure of (E)-1-(((2-amino-4,5-dimethylphenyl)iminio)methyl)naphthalen-2-olate, C19H18N2O
- Crystal structure of N-(acridin-9-yl)-2-(4-methylpiperidin-1-yl) acetamide monohydrate, C21H25N3O2
- The crystal structure of dichlorido-bis(3-methyl-3-imidazolium-1-ylpropionato-κ2 O,O′)-zinc(II), C14H20Cl2N4O4Zn
- The crystal structure of 2,8-diethyl-1,3,7,9-tetramethyl-4λ4,5λ4-spiro[dipyrrolo[1,2-c:2′,1′-f][1,3,2]diazaborinine-5,2′-naphtho[1,8-de][1,3,2]dioxaborinine], C25H29BN2O2
- The crystal structure of 5-tert-butyl-2-(5-tert-butyl-3-iodo-benzofuran-2-yl)-3-iodobenzofuran, C24H24I2O2
- Synthesis and crystal structure of methyl 2-{[4-(4-cyclopropyl-1-naphthyl)-4H-1,2,4-triazole-3-yl]thio} acetate, C18H17N3O2S
- The crystal structure of n-propylammonium bis(2,3-dimethylbutane-2,3-diolato)borate-boric acid (1/1), [C3H10N][C12H24BO4]·B(OH)3
- Crystal structure of methyl 1-(2-bromophenyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate, C19H17BrN2O2
- Crystal structure of (4-bromobenzyl)triphenylphosphonium bromide ethanol solvate, C52H48Br4OP2
- The crystal structure of unsymmetrical BOPHY C26H27BN4
- The crystal structure of Tb3B5O11(OH)2
- The crystal structure of (Z)-4-ethyl-2-((4-ethyl-3,5-dimethyl-1H-pyrrol-2-yl)methylene)-3,5-dimethyl-2H-pyrrol-1-ium 2,2'-spirobi[naphtho[1,8-de][1,3,2]dioxaborinin]-2-uide, C37H37BN2O4
- Crystal structure of bis(methylammonium) hexadecaselenidopalladate(II), (CH3NH3)2PdSe16
- The crystal structure of (2-diphenylphosphanylphenyl) 2-[7-(dimethylamino)-2-oxochromen-4-yl]acetate, C31H26NO4P
- Crystal structure of (E)-6-(4-ethylpiperazin-1-yl)-2-(3-fluorobenzylidene)-3,4-dihydronaphthalen-1(2H)-one, C23H25FN2O
- The structure of RUB-56, (C6H16N)8 [Si32O64(OH)8]·32 H2O, a hydrous layer silicate (2D-zeolite) that contains microporous levyne-type silicate layers
- Crystal structure of 4-amino-3,5-dibromobenzonitrile, C7H4Br2N2
- Crystal structure of 2-(naphthalen-1-yl)ethyl 2-acetoxybenzoate, C21H18O4
- Single-crystal structure determination of Tm3B12O19(OH)7
- Crystal structure determination of NdB3.6O7
- The crystal structure of NdB6O8(OH)5·H3BO3
- Crystal structure of 2-(5-ethylpyridin-2-yl)ethyl 2-(6-methoxynaphthalen-2-yl)propanoate, C23H25NO3
- Crystal structure of N-(1-(3,4-dimethoxyphenyl)-2-methylpropyl)aniline, C18H23NO2
- Crystal structure of Ba6Cd12Mn4SiF48
- Synthesis and crystal structure of 5-fluoro-1-methyl-2-oxo-3-(2-oxochroman-4-yl)indolin-3-yl acetate, C20H16FNO5
- The crystal structure of 6-methacryloylbenzo[d][1,3]dioxol-5-yl 4-nitrobenzenesulfonate, C17H13NO8S
- Crystal structure of ethyl 2-(3-benzyl-4-oxo-3,4-dihydrophthalazin-1-yl)- 2,2-difluoroacetate, C19H16F2N2O3
- The crystal structure of tetrakis(μ 2-(1H-benzimidazole-2-methoxo-κ2 N,O:O:O)-(n-butanol-κO)-chlorido)-tetranickel(II), C48H68Cl4N8O8Ni4
- Synthesis and crystal structure of trans-tetraaqua-bis((1-((7-hydroxy-3-(4-methoxy-3-sulfonatophenyl)-4-oxo-4H-chromen-8-yl)methyl)piperidin-1-ium-4-carbonyl)oxy-κO)zinc(II)hexahydrate, C46H64N2O28S2Zn
- The crystal structure of 1-(4-carboxybutyl)-3-methyl-1H-imidazol-3-ium hexafluoridophosphate, C9H15F6N2O2P
- Crystal structure of 1-(4-chlorophenyl)-4-(2-furoyl)-3-phenyl-1H-pyrazol-5-ol, C20H13ClN2O3
- Crystal structure of dimethyl (R)-2-(3-(1-phenylethyl)thioureido)-[1,1′-biphenyl]-4,4′-dicarboxylate, C25H24N2O4S
- The crystal structure of 1-(3-carboxypropyl)-1H-imidazole-3-oxide, C7H10N2O3
- Synthesis and crystal structure of dimethyl 4,4′-(propane-1,3-diylbis(oxy))dibenzoate, C19H20O6
- Crystal structure of methyl-1-(p-tolyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate, C20H20N2O2
- The crystal structure of 1-(1-adamantan-1-yl)ethyl-3-(3-methoxyphenyl)thiourea, C20H28N2OS
- The crystal structure of N,N′-carbonylbis(2,6-difluorobenzamide), C15H8F4N2O3
Articles in the same Issue
- Frontmatter
- New Crystal Structures
- The crystal structure of tris((Z)-2-hydroxy-N-((E)-pyridin-2-ylmethylene)benzohydrazonato-k2O,N)europium(III), C39H30N9O6Eu
- Crystal structure of (E)-3-(benzylideneamino)-2-phenylthiazolidin-4-one, C16H14N2OS
- The crystal structure of (E)-4-fluoro-N′-(1-(o-tolyl)ethylidene)benzohydrazide, C16H15FN2O
- Crystal structure of (6-chloropyridin-3-yl)methyl 2-(6-methoxynaphthalen-2-yl)propanoate, C20H18ClNO3
- Crystal structure of methyl 3-methoxy-4-(2-methoxy-2-oxoethoxy)benzoate, C12H14O6
- The crystal structure of bis[(4-methoxyphenyl)(picolinoyl)amido-κ2 N:N′]copper(II), C26H22CuN4O4
- The crystal structure of poly[di(μ2-aqua)-diaqua-bis(3-aminopyridine-4-carboxylate-κ2 O: O′)-tetra(μ2-3-aminopyridine-4-carboxylate-κ2 O: O′)-dineodymium(III), [Nd2(C6H5N2O2)6(H2O)4] n
- The crystal structure of t-butyl 7-[3-(4-fluorophenyl)-1-(propan-2-yl)-1H-indol-2-yl]-3,5-dihydroxyhept-6-enoate, C28H34FNO4
- Crystal structure of catena-poly[(benzylamine-κ1 N)-(sorbato-κ1 O)-(μ2-sorbato-κ2 O,O′)-copper(II), C19H23CuNO4
- Crystal structure of (4-(2-chlorophenyl)-1H-pyrrol-3-yl)(ferrocenyl) methanone, C21H16ClFeNO
- The crystal structure of N-[4-(4-bromophenyl)-1,3-thiazol-2-yl]-3-(2-methylphenyl)-2-sulfanylprop-2-enamide hydrate, C19H17BrN2O2S2
- The crystal structure of N′-{5-[2-(2,6-dimethylphenoxy) acetamido]-4-hydroxy-1,6-diphenylhexan-2-yl}-3-methyl-2-(2-oxo-1,3-diazinan-1-yl)butanamide hydrate
- Crystal structure of 2-(naphthalen-1-yl)ethyl 2-(6-methoxynaphthalen-2-yl)propanoate, C26H24O3
- Crystal structure of naphthalen-1-ylmethyl 2-(6-methoxynaphthalen-2-yl)propanoate, C25H22O3
- Crystal structure of poly[diaqua- (μ4-5-(1H-1,2,4-triazol-1-yl)benzene-1,3-dicarboxylato-κ5N:O,O’:O’’:O’’’)calcium(II), C10H9CaN3O6
- Crystal structure of (E)-N′-(4-((E)-3-(dimethylamino)acryloyl)-3-hydroxyphenyl)-N, N-dimethylformimidamide, C14H19N3O2
- Crystal structure of (E)-3-(dimethylamino)-1-(2-hydroxy-4,6-dimethoxyphenyl)prop-2-en-1-one, C13H17NO4
- Crystal structure of (2-chloropyridin-3-yl)methyl-2-(6-methoxynaphthalen-2-yl)propanoate, C20H18ClNO3
- The crystal structure of diethyl 4-(3,4-dimethylphenyl)-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate, C21H27NO4
- Crystal structure of (8R,9S,10R,13S,14S,17S)-17-hydroxy-10,13-dimethyl-17-((4-(2-phenylpropyl)phenyl)ethynyl)-1,2,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-3H-cyclopenta[a]phenanthren-3-one, C36H42O2
- Synthesis and crystal structure of 4-(4-cyclopropylnaphthalen-1-yl)-2,4-dihydro-3H-1,2,4-triazole-3-thione, C15H13N3S
- Crystal structure of catena-poly[aqua-(2,6-di-(2-pyridyl)-pyridine-κ3 N,N′, N″)(μ2-1,4-naphthalene dicarboxylato-κ2 O,O′)nickel(II)], C27H19NiN3O5
- Crystal structure of 3-(diphenylphosphoryl)-3-hydroxy-1-phenylpropan-1-one, C21H19O3P
- The crystal structure of R,S-{N-[(2-oxidonaphthalen-1-yl)methylidene]phenylglycinato}divinylsilicon, C23H19NO3Si
- The crystal structure of 1,2,4-tris(bromomethyl)benzene, C9H9Br3
- Crystal structure of chlorido-[4-(pyridin-2-yl)benzaldehyde-κ2 N,C]-(diethylamine-κ1 N)platinum(II), C16H18ClN2OPt
- Crystal structure of 3-(methoxycarbonyl)-1-(4-methoxyphenyl)-2,3,4,9- tetrahydro-1H-pyrido[3,4-b]indol-2-ium chloride hydrate, C40H48Cl2N4O9
- The crystal structure of 1-(2-chlorobenzyl)-3-(3-chlorophenyl)urea, C14H12Cl2N2O
- Hydrothermal synthesis and crystal structure of aqua-tris(4-acetamidobenzoato-κ2 O,O′)-(1,10-phenanthroline-κ2 N,N′)terbium(III) hydrate C39H36N5O11Tb
- The crystal structure of zwitterionic 3-aminoisonicotinic acid, C6H6N2O2
- The crystal structure of bis{[monoaqua-μ2-4-[(pyridine-4-carbonyl)-amino]-phthalato-κ3 N:O,O′-(2,2′-bipyridine κ2 N,N′)copper(II)]}decahydrate, C48H56N8O22Cu2
- Crystal structure of poly[μ10-4,4′-methylene-bis(oxy)benzoatodipotassium], C15H10K2O6
- The crystal structure of catena-poly[[tetraaqua[(μ2-1,4-di(4-methyl-1-imidazolyl)benzene] cobalt(II)]bis(formate)], C16H24CoN4O8
- The crystal structure of (E)-2-chloro-5-((2-(nitromethylene)imidazolidin-1-yl)methyl)pyridine, C10H11ClN4O2
- The crystal structure of (E)-1-(((2-amino-4,5-dimethylphenyl)iminio)methyl)naphthalen-2-olate, C19H18N2O
- Crystal structure of N-(acridin-9-yl)-2-(4-methylpiperidin-1-yl) acetamide monohydrate, C21H25N3O2
- The crystal structure of dichlorido-bis(3-methyl-3-imidazolium-1-ylpropionato-κ2 O,O′)-zinc(II), C14H20Cl2N4O4Zn
- The crystal structure of 2,8-diethyl-1,3,7,9-tetramethyl-4λ4,5λ4-spiro[dipyrrolo[1,2-c:2′,1′-f][1,3,2]diazaborinine-5,2′-naphtho[1,8-de][1,3,2]dioxaborinine], C25H29BN2O2
- The crystal structure of 5-tert-butyl-2-(5-tert-butyl-3-iodo-benzofuran-2-yl)-3-iodobenzofuran, C24H24I2O2
- Synthesis and crystal structure of methyl 2-{[4-(4-cyclopropyl-1-naphthyl)-4H-1,2,4-triazole-3-yl]thio} acetate, C18H17N3O2S
- The crystal structure of n-propylammonium bis(2,3-dimethylbutane-2,3-diolato)borate-boric acid (1/1), [C3H10N][C12H24BO4]·B(OH)3
- Crystal structure of methyl 1-(2-bromophenyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate, C19H17BrN2O2
- Crystal structure of (4-bromobenzyl)triphenylphosphonium bromide ethanol solvate, C52H48Br4OP2
- The crystal structure of unsymmetrical BOPHY C26H27BN4
- The crystal structure of Tb3B5O11(OH)2
- The crystal structure of (Z)-4-ethyl-2-((4-ethyl-3,5-dimethyl-1H-pyrrol-2-yl)methylene)-3,5-dimethyl-2H-pyrrol-1-ium 2,2'-spirobi[naphtho[1,8-de][1,3,2]dioxaborinin]-2-uide, C37H37BN2O4
- Crystal structure of bis(methylammonium) hexadecaselenidopalladate(II), (CH3NH3)2PdSe16
- The crystal structure of (2-diphenylphosphanylphenyl) 2-[7-(dimethylamino)-2-oxochromen-4-yl]acetate, C31H26NO4P
- Crystal structure of (E)-6-(4-ethylpiperazin-1-yl)-2-(3-fluorobenzylidene)-3,4-dihydronaphthalen-1(2H)-one, C23H25FN2O
- The structure of RUB-56, (C6H16N)8 [Si32O64(OH)8]·32 H2O, a hydrous layer silicate (2D-zeolite) that contains microporous levyne-type silicate layers
- Crystal structure of 4-amino-3,5-dibromobenzonitrile, C7H4Br2N2
- Crystal structure of 2-(naphthalen-1-yl)ethyl 2-acetoxybenzoate, C21H18O4
- Single-crystal structure determination of Tm3B12O19(OH)7
- Crystal structure determination of NdB3.6O7
- The crystal structure of NdB6O8(OH)5·H3BO3
- Crystal structure of 2-(5-ethylpyridin-2-yl)ethyl 2-(6-methoxynaphthalen-2-yl)propanoate, C23H25NO3
- Crystal structure of N-(1-(3,4-dimethoxyphenyl)-2-methylpropyl)aniline, C18H23NO2
- Crystal structure of Ba6Cd12Mn4SiF48
- Synthesis and crystal structure of 5-fluoro-1-methyl-2-oxo-3-(2-oxochroman-4-yl)indolin-3-yl acetate, C20H16FNO5
- The crystal structure of 6-methacryloylbenzo[d][1,3]dioxol-5-yl 4-nitrobenzenesulfonate, C17H13NO8S
- Crystal structure of ethyl 2-(3-benzyl-4-oxo-3,4-dihydrophthalazin-1-yl)- 2,2-difluoroacetate, C19H16F2N2O3
- The crystal structure of tetrakis(μ 2-(1H-benzimidazole-2-methoxo-κ2 N,O:O:O)-(n-butanol-κO)-chlorido)-tetranickel(II), C48H68Cl4N8O8Ni4
- Synthesis and crystal structure of trans-tetraaqua-bis((1-((7-hydroxy-3-(4-methoxy-3-sulfonatophenyl)-4-oxo-4H-chromen-8-yl)methyl)piperidin-1-ium-4-carbonyl)oxy-κO)zinc(II)hexahydrate, C46H64N2O28S2Zn
- The crystal structure of 1-(4-carboxybutyl)-3-methyl-1H-imidazol-3-ium hexafluoridophosphate, C9H15F6N2O2P
- Crystal structure of 1-(4-chlorophenyl)-4-(2-furoyl)-3-phenyl-1H-pyrazol-5-ol, C20H13ClN2O3
- Crystal structure of dimethyl (R)-2-(3-(1-phenylethyl)thioureido)-[1,1′-biphenyl]-4,4′-dicarboxylate, C25H24N2O4S
- The crystal structure of 1-(3-carboxypropyl)-1H-imidazole-3-oxide, C7H10N2O3
- Synthesis and crystal structure of dimethyl 4,4′-(propane-1,3-diylbis(oxy))dibenzoate, C19H20O6
- Crystal structure of methyl-1-(p-tolyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate, C20H20N2O2
- The crystal structure of 1-(1-adamantan-1-yl)ethyl-3-(3-methoxyphenyl)thiourea, C20H28N2OS
- The crystal structure of N,N′-carbonylbis(2,6-difluorobenzamide), C15H8F4N2O3

